Thermophiles in Astrobiology and Biotechnology: Difference between revisions
Pmedina0098 (talk | contribs) |
Pmedina0098 (talk | contribs) |
||
| (26 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
{{Uncurated}} | {{Uncurated}} | ||
A thermophile is an organism that thrives at relatively high temperatures, between 45 and 122°C. Thermophiles are found in a | A thermophile is an organism that thrives at relatively high temperatures, between 45 and 122°C. Thermophiles are found in a number of marine and terrestrial geothermally-heated habitats including shallow terrestrial hot springs, hydrothermal vent systems, sediment from volcanic islands, and deep sea hydrothermal vents. As prerequisite for their survival, thermophiles contain enzymes that can function at high temperatures. Investigating the DNA and protein stability of thermophiles is important because they can reveal what extraterrestrial life may look like and can give insights in biotechnology. | ||
==Introduction== | ==Introduction== | ||
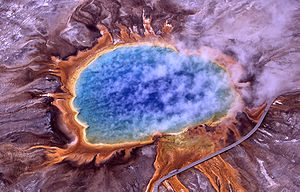
<br>The National Center for Biotechnology Inforation (NCBI) Microbial Genome Project Database uses five terms to categorize the temperature range an organism grows at, where [http://en.wikipedia.org/wiki/Cryophilic cryophilic] refers to –30° to –2°C, [http://en.wikipedia.org/wiki/Cryophilic psychrophilic] refers to –1° to +10°C, [http://en.wikipedia.org/wiki/Mesophile mesophilic] refers to +11° to +45°C, [http://en.wikipedia.org/wiki/Thermophile thermophilic] refers to +46° to 75°C, and [http://en.wikipedia.org/wiki/Hyperthermophilic hyperthermophilic] refers to above +75°C. [[Image:300px-Grand_prismatic_spring.jpg|thumb|300px|right|Figure 1. Aerial view of Grand Prismatic Spring in Yellowstone National Park. Thermophiles produce some of the bright colors seen here. Image from examiner.com]] | <br>The National Center for Biotechnology Inforation (NCBI) Microbial Genome Project Database uses five terms to categorize the temperature range an organism grows at, where [http://en.wikipedia.org/wiki/Cryophilic cryophilic] refers to –30° to –2°C, [http://en.wikipedia.org/wiki/Cryophilic psychrophilic] refers to –1° to +10°C, [http://en.wikipedia.org/wiki/Mesophile mesophilic] refers to +11° to +45°C, [http://en.wikipedia.org/wiki/Thermophile thermophilic] refers to +46° to 75°C, and [http://en.wikipedia.org/wiki/Hyperthermophilic hyperthermophilic] refers to above +75°C. [[Image:300px-Grand_prismatic_spring.jpg|thumb|300px|right|Figure 1. Aerial view of Grand Prismatic Spring in Yellowstone National Park. Thermophiles produce some of the bright colors seen here. Image from examiner.com]] Thermophiles are found in geothermally heated regions of the Earth like deep-sea [http://en.wikipedia.org/wiki/Hydrothermal_vent hydrothermal vents] and the hot springs of [http://en.wikipedia.org/wiki/Yellowstone_National_Park Yellowstone National Park] (Figure 1). The investigation of thermophilic structure and chemistry poses very promising and intriguing contributions to the scientific community. For one, some of the enzymes used in molecular biology, like [http://en.wikipedia.org/wiki/DNA_polymerase DNA polymerases], have derived from investigating heat-stable enzymes. Perhaps the quintessential example of a successful biotechnological application of thermozymes is the use of Taq polymerase, isolated from <i>Thermus aquaticus</i> <sup>[http://jb.asm.org/content/98/1/289.short 6]</sup>, for PCR. | ||
<br> | |||
In addition, astrobiologists look to understand the structural and genomic correlates of hyperthermostability to give indication to what life may look like on planets hotter than ours. Also, astrobiologists, including researchers from NASA, suggest that hot springs all over the world provide some of the best "doorways into early Earth." <sup>[http://www.ncbi.nlm.nih.gov/pmc/articles/PMC545586/ 7]</sup> Many scientists believe that life might have begun roughly 4 billion years ago in high temperature environments and that the first organisms might therefore be thermophiles. Not only does this give insight into the origin of life on Earth, but opens up a new realm of possibilities for life elsewhere in the universe.<sup>[http://www.ncbi.nlm.nih.gov/pmc/articles/PMC545586/ 7]</sup><br> | |||
<br>Over the last 20 years, researchers have begun to discover key physical adaptations of proteins and DNA that allow thermophiles to remain functional and alive at high temperatures. First, increasing the number of [http://en.wikipedia.org/wiki/Salt_bridge_(protein_and_supramolecular) salt bridges] is a driving force for enhancement of the thermotolerance of proteins from hyperthermophilic microorganisms.<sup>[http://www.sciencedirect.com/science/article/pii/S0968000401019181 2]</sup> Second, research suggests that the replacement of polar noncharged residues by charged ones constitutes a major stabilization mechanisms in the proteins of hyperthermophilic organisms.<sup>[http://www.sciencedirect.com/science/article/pii/S0022283603013147 3]</sup> Third, disulfide bond abundance has been observed increases as a function of the maxiumum growth temperature of an organisms increases.<sup>[http://www.plosbiology.org/article/info%3Adoi%2F10.1371%2Fjournal.pbio.0030309 14]</sup> Fourth, thermophilic protein sequences are more likely than their mesophilic homologs to have deletions in exposed [http://en.wikipedia.org/wiki/Stem-loop loop regions]. Lastly, there is evidence that the [http://en.wikipedia.org/wiki/GC-content guanine-cytosine] (GC) content levels of the coding/non-coding regions of certain genes are highly likely to be correlated with the temperature range conditions of prokaryotic organisms.<br> | |||
<br> | |||
<br> <br> | <br> <br> | ||
==Protein Stabilization== | ==Protein Stabilization== | ||
High temperatures can often denature vital enzymes and proteins. | High temperatures can often denature vital enzymes and proteins. Heating affects the secondary structure of proteins, causing changes in the shape of the molecule. Specifically, heat disrupts hydrogen bonds and non-polar hydrophobic interactions. This occurs because heat increases the kinetic energy and cause the molecules to vibrate so rapidly and violently that the bonds are disrupted. Changing a proteins shape can alter its natural function thereby disrupting vital processes like metabolism. In order to withstand hot temperatures, thermophilic proteins can exhibit higher core hydrophobicity<sup>[http://onlinelibrary.wiley.com/doi/10.1002/pro.5560021007/ 9]</sup>, greater numbers of ionic interactions<sup>[http://www.pnas.org/content/95/21/12300.short 10]</sup>, increased packing density<sup>[http://pubs.acs.org/doi/abs/10.1021/bi9705321 11]</sup>, additional networks of hydrogen bonds<sup>[http://www.sciencedirect.com/science/article/pii/S0959440X98800948 12]</sup>, decreased lengths of surface loops<sup>[http://www.sciencedirect.com/science/article/pii/S0022283699928899 4]</sup>, stabilization by heat stable chaperones<sup>[http://www.nature.com/nsmb/journal/v12/n10/full/nsmb993.html 13]</sup>, and increase in disulfide bond formation<sup>[http://www.plosbiology.org/article/info%3Adoi%2F10.1371%2Fjournal.pbio.0030309 14]</sup> and a general shortening of length<sup>[http://www.pasteur.fr/~tekaia/tekaia.GENE.2002.pdf 15]</sup>. | ||
<br> | <br> | ||
| Line 19: | Line 19: | ||
====Salt Bridges==== | ====Salt Bridges==== | ||
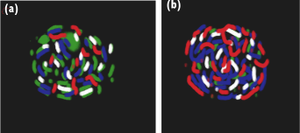
[[Image:salt_bridge_paint.png|thumb|300px|right|Figure 2. Comparison of the capsid surface residues in lumazine synthase from (a) <i>Bacillus subtilis</i> and (b) the hyperthermophile <i>Aquifex aeolicus</i>]] [[Image:salt_bridge_table.png|thumb|300px|right|Table 1. [http://www.sciencedirect.com/science/article/pii/S0968000401019181 Number of salt bridges in proteins from thermo- and hyper thermophilic organisms] (see Table 1 in Karshikoff and Ladenstein 2001)]] The optimization of [http://www.cliffsnotes.com/study_guide/Electrostatic-and-van-der-Waals-Interactions.topicArticleId-24998,articleId-24940.html electrostatic interactions] by increasing the number of salt bridges is a driving force for enhancement of the thermotolerance of proteins from hyperthermophilic microorganisms. <sup>[http://www.sciencedirect.com/science/article/pii/S0968000401019181 2]</sup> This trend is less evident in thermophilic organisms and absent from mesophile-derived proteins | [[Image:salt_bridge_paint.png|thumb|300px|right|Figure 2. Comparison of the capsid surface residues in lumazine synthase from (a) <i>Bacillus subtilis</i> and (b) the hyperthermophile <i>Aquifex aeolicus</i>]] [[Image:salt_bridge_table.png|thumb|300px|right|Table 1. [http://www.sciencedirect.com/science/article/pii/S0968000401019181 Number of salt bridges in proteins from thermo- and hyper thermophilic organisms] (see Table 1 in Karshikoff and Ladenstein 2001)]] A salt bridge is a combination of two [http://en.wikipedia.org/wiki/Noncovalent_bonding noncovalent] interactions: hydrogen bonding and electrostatic interactions. The optimization of [http://www.cliffsnotes.com/study_guide/Electrostatic-and-van-der-Waals-Interactions.topicArticleId-24998,articleId-24940.html electrostatic interactions] by increasing the number of salt bridges is a driving force for enhancement of the thermotolerance of proteins from hyperthermophilic microorganisms. <sup>[http://www.sciencedirect.com/science/article/pii/S0968000401019181 2]</sup> This trend is less evident in thermophilic organisms and absent from mesophile-derived proteins. Salt bridges often occur between groups distant in the protein sequence and form [http://en.wikipedia.org/wiki/Cross-link cross-links] that stabilize [http://en.wikipedia.org/wiki/Protein_structure tertiary structure]. This interaction can increase the [http://en.wikipedia.org/wiki/Energy_barrier kinetic barrier] towards thermal inactivation or thermal unfolding and, thus, prevents proteins from denaturing at high temperatures. | ||
<br>Table 1 shows the number of salt bridges in select thermo- and hyperthermophilic organisms. N<sub>s</sub> indicates the number of salt bridges, N<sub>r</sub> represents the number of salt bridges statistically expected for that protein structure, and T<sub>opt</sub> represents the temperature of optimal growth for the protein. Proteins from hyperthermophilic organisms are characterized by an increased number of ion pairs with respect to the statistical expectance and/or the number of ion-pairs in their mesophilic counterparts. This finding suggests that electrostatic interactions are a principal factor responsible for the elevation of the melting temperature of proteins from hyperthermophilic organisms. | <br>Table 1 shows the number of salt bridges in select thermo- and hyperthermophilic organisms. N<sub>s</sub> indicates the number of salt bridges, N<sub>r</sub> represents the number of salt bridges statistically expected for that protein structure, and T<sub>opt</sub> represents the temperature of optimal growth for the protein. Proteins from hyperthermophilic organisms are characterized by an increased number of ion pairs with respect to the statistical expectance and/or the number of ion-pairs in their mesophilic counterparts. This finding suggests that electrostatic interactions are a principal factor responsible for the elevation of the melting temperature of proteins from hyperthermophilic organisms. | ||
<br> | <br><br> | ||
The figure and table | The figure and table to the right show that charged residues in the enzyme from <i>Aquifex aeolicus</i> replace most of the polar residues in the <i>Bacillus subtilis</i> enzyme. Specifically, the number of ion pairs in the protein from <i>Aquifex</i> is increased by >90%. Furthermore, melting point assessments are often employed in thermostability studies in order to examine the effect of structural changes. In this case, the corresponding change in melting temperature when <i>B. subtilis</i> to <i>A. aeolicus</i> was about 27°C. <sup>[http://www.sciencedirect.com/science/article/pii/S0968000401019181 2]</sup> | ||
<br>Molecular dynamics calculations on a prototypical ion-pair model system have suggested that a sizeable energy barrier exists for the solvation of a salt bridge and that the height of this barrier increases with temperature. Interestingly, a similar barrier is not seen with [http://en.wikipedia.org/wiki/Isostere isosteric] hydrophobic groups. Thus, the heightened kinetic barrier towards protein unfolding is one of the mechanisms thermophiles can employ to stabilize their proteins and prevent them from denaturing. <br> | <br><br> | ||
Molecular dynamics calculations on a prototypical ion-pair model system have suggested that a sizeable energy barrier exists for the solvation of a salt bridge and that the height of this barrier increases with temperature. Interestingly, a similar barrier is not seen with [http://en.wikipedia.org/wiki/Isostere isosteric] hydrophobic groups. Thus, the heightened kinetic barrier towards protein unfolding is one of the mechanisms thermophiles can employ to stabilize their proteins and prevent them from denaturing. <br> | |||
<br><br> | <br><br> | ||
====Polar Charged Residues==== | ====Polar Charged Residues==== | ||
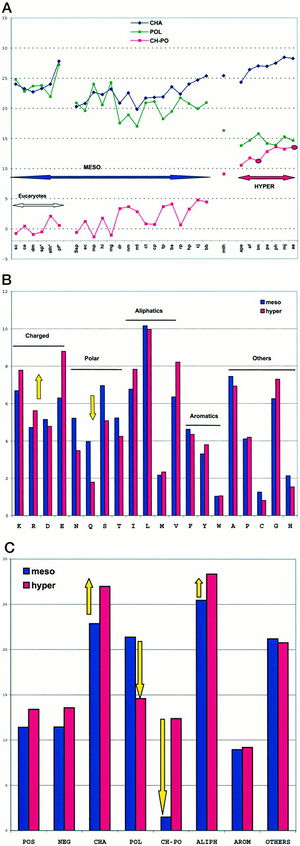
[[Image:polar charged residues.jpg|thumb|300px|right|Figure 3. Amino acid content variations within 30 genomes. Cambillau and Claverie 2000.]] The charged polar amino acid content is a genomic signature that can give strong indications to an organisms growth environment. The advantage of polar amino acids comes from the increased stability of coulombic interactions with increasing temperatures. Therefore, in most structures of hyperthermophilic proteins, the existence of long chains of ion pairs provide cooperative stabilization. <sup>[http://www.sciencedirect.com/science/article/pii/S0022283603013147 3]</sup> This finding implies the existence of global structural features associated with hyperthermostability common to thermophilic Bacteria and Achaea as described below. | [[Image:polar charged residues.jpg|thumb|300px|right|Figure 3. Amino acid content variations within 30 genomes. Cambillau and Claverie 2000.]] The charged polar amino acid content is a genomic signature that can give strong indications to an organisms growth environment. The advantage of polar amino acids comes from the increased stability of [http://en.wikipedia.org/wiki/Coulomb's_law coulombic interactions] with increasing temperatures. Therefore, in most structures of hyperthermophilic proteins, the existence of long chains of ion pairs provide cooperative stabilization. <sup>[http://www.sciencedirect.com/science/article/pii/S0022283603013147 3]</sup> This finding implies the existence of global structural features associated with hyperthermostability common to thermophilic Bacteria and Achaea as described below. | ||
<br> | <br> | ||
The replacement of polar noncharged residues by charged ones constitutes a major stabilization mechanism in the proteins of hyperthermophilic organisms. Residue changes allow the stabilization of proteins through ion bonds. A stronger intramolecular interaction between the protein and itself decreases the effect of an intermolecular force such as heat. In other words, more heat is required to denature the protein if there are stronger intramolecular interactions taking place. The proteome analysis in amino acid classes indicated that hyperthermophilicity is characterized by a sharp increase of charged residues, Lys and Glu, at the expense of polar noncharged residues, mainly Gln (Figure 3). Figure 3A is a plot of the sum of the percentages of charged amino acids (Lys, Arg, Asp, Glu; CHA, blue), polar noncharged amino acids (Asn, Gln, Ser, Thr; POL, green), and of the difference of the two values (CH-POL, red). The mesophiles and hyperthermophiles are identified by MESO and HYPER, respectively. Figure 3B is a plot of the percentages of the various amino acids in mesophiles (blue) and hyperthermophiles (red). Figure 3C is a plot of the sum of percentages of the various amino acid classes in mesophiles (blue) and hyperthermophiles (red). A threshold is observed around 10% for extremeophiles Ch-Po values, while a higher limit of 5% characterizes mesophiles. <sup>[http://www.jbc.org/content/275/42/32383.full 1]</sup>Cambillau and Claverie suggest from this data that the difference between charged and polar noncharged amino acids (Ch-Po) is the best indicator of an organism’s lifestyle.<sup>[http://www.jbc.org/content/275/42/32383.full 1]</sup><br> | The replacement of polar noncharged residues by charged ones constitutes a major stabilization mechanism in the proteins of hyperthermophilic organisms. Residue changes allow the stabilization of proteins through ion bonds. A stronger intramolecular interaction between the protein and itself decreases the effect of an intermolecular force such as heat. In other words, more heat is required to denature the protein if there are stronger intramolecular interactions taking place. The proteome analysis in amino acid classes indicated that hyperthermophilicity is characterized by a sharp increase of charged residues, Lys and Glu, at the expense of polar noncharged residues, mainly Gln (Figure 3). Figure 3A is a plot of the sum of the percentages of charged amino acids (Lys, Arg, Asp, Glu; CHA, blue), polar noncharged amino acids (Asn, Gln, Ser, Thr; POL, green), and of the difference of the two values (CH-POL, red). The mesophiles and hyperthermophiles are identified by MESO and HYPER, respectively. Figure 3B is a plot of the percentages of the various amino acids in mesophiles (blue) and hyperthermophiles (red). Figure 3C is a plot of the sum of percentages of the various amino acid classes in mesophiles (blue) and hyperthermophiles (red). A threshold is observed around 10% for extremeophiles Ch-Po values, while a higher limit of 5% characterizes mesophiles. <sup>[http://www.jbc.org/content/275/42/32383.full 1]</sup>Cambillau and Claverie suggest from this data that the difference between charged and polar noncharged amino acids (Ch-Po) is the best indicator of an organism’s lifestyle.<sup>[http://www.jbc.org/content/275/42/32383.full 1]</sup><br> | ||
| Line 35: | Line 36: | ||
====Disulfide Bonding==== | ====Disulfide Bonding==== | ||
Structural disulfide bonds are a covalent tertiary interaction in extracellular and compartmentalized proteins, acting to stabilize a folded protein structure. The disulfide bond stabilizes the folded form of a protein in several ways. First, it holds two | Structural disulfide bonds are a covalent tertiary interaction in extracellular and compartmentalized proteins, acting to stabilize a folded protein structure. The disulfide bond stabilizes the folded form of a protein in several ways. First, it holds two portions of the protein together, baising the protein towards the folded topology. Second, the disulfide bond may form the nucleus of a hydrophobic core of the folded protein, i.e., local hydrophobic residues may condense around the disulfide bond and onto each other through hydrophobic interactions. Third, the disulfide bond link two segments of the protein chain, the disulfide bond increases the effective local concentration of protein residues and lowers the effective local concentration of water molecules.<sup>[http://en.wikipedia.org/wiki/Disulfide_bond 16]</sup> | ||
<br><br> | |||
The specific distribution of disulfide bonds observed across prokaryotic genomes suggests specialization in strategies used by organisms to stabilize their proteins. Trends in pairwise amino acid proximities were measure for proteins from 199 distinct prokaryotic genomes, and close cysteine-cysteine pairings were interpreted as likely specific disulfide bonds in these organisms.<sup>[http://www.plosbiology.org/article/info%3Adoi%2F10.1371%2Fjournal.pbio.0030309 14]</sup> Thermophiles exhibited a pronounced bias in the spatial proximity of cysteine-cysteine residues, supporting a role for disulfide bonds in these organisms. The predicted disulfide abundance (expressed as a proximity score for cysteine-cysteine pairs) is shown in Figure 3 as a function of the maximum growth temperature of each organism. Here, a plot of log ratios of | The specific distribution of disulfide bonds observed across prokaryotic genomes suggests specialization in strategies used by organisms to stabilize their proteins. Trends in pairwise amino acid proximities were measure for proteins from 199 distinct prokaryotic genomes, and close cysteine-cysteine pairings were interpreted as likely specific disulfide bonds in these organisms.<sup>[http://www.plosbiology.org/article/info%3Adoi%2F10.1371%2Fjournal.pbio.0030309 14]</sup> Thermophiles exhibited a pronounced bias in the spatial proximity of cysteine-cysteine residues, supporting a role for disulfide bonds in these organisms. The predicted disulfide abundance (expressed as a proximity score for cysteine-cysteine pairs) is shown in Figure 3 as a function of the maximum growth temperature of each organism. Here, a plot of log ratios of disulfide richness is identified in thermophiles, both archaeal and bacterial. | ||
<br><br> | |||
[[Image:disulfide.jpeg|thumb|300px|left|Figure 3. Correspondence of Growth Temperature and Disulfide Richness. Organisms are classified by color and symbol shape according to the following scheme: mesophiles that have been suggested to have extremophilic qualities (blue); mesophiles with no suggested extremophilic qualities (grey); sulfur-reducing bacteria and archaea (yellow); methanogenic bacteria and archaea (green), non-methanogenic/non-sulfur-reducing thermophiles (red); genomes that contain a PDO protein (triangle)l and genomes that do not contain a PDO protein (circle). Beeby <i>et al.</i> 2005.]] In addition to thermophilic prokaryotes, certain other organisms appear to have measurably elevated degrees of disulfide bonding. These include some halophiles, alkalophiles, acidophiles, and radiation-tolerant organisms. This trend suggests that disulfide bonds might serve generally to stabilize proteins in a variety of extreme environments. | [[Image:disulfide.jpeg|thumb|300px|left|Figure 3. Correspondence of Growth Temperature and Disulfide Richness. Organisms are classified by color and symbol shape according to the following scheme: mesophiles that have been suggested to have extremophilic qualities (blue); mesophiles with no suggested extremophilic qualities (grey); sulfur-reducing bacteria and archaea (yellow); methanogenic bacteria and archaea (green), non-methanogenic/non-sulfur-reducing thermophiles (red); genomes that contain a PDO protein (triangle)l and genomes that do not contain a PDO protein (circle). Beeby <i>et al.</i> 2005.]] In addition to thermophilic prokaryotes, certain other organisms appear to have measurably elevated degrees of disulfide bonding. These include some halophiles, alkalophiles, acidophiles, and radiation-tolerant organisms. This trend suggests that disulfide bonds might serve generally to stabilize proteins in a variety of extreme environments. | ||
| Line 47: | Line 48: | ||
<br><br> | <br><br> | ||
In one noteable example, Usher <i>et al.</i> compared the structures of the CheY protein from <i>Thermotoga maritima</i> and <i> Escherichia coli</i>, and found no increase in ion pairs, ion-pair networks or hydrogen bonding.<sup>[http://onlinelibrary.wiley.com/doi/10.1002/pro.5560070221/abstract 20]</sup> Rather, they observed a shortening of the N and C termini of the thermostable protein, truncation of one of its loops and an increase in proline residues, all [http://en.wikipedia.org/wiki/Entropic_force entropic] factors. On the low end of the temperature scale, the inverse effect has been observed. From comparisons of structure from mesophilic or thermophilic <i>versus</i> psychrophilic organisms the psychrophilic proteins are found to have insertions in exposed loop regions.<sup>[http://www.pnas.org/content/89/11/5142 21]</sup> | In one noteable example, Usher <i>et al.</i> compared the structures of the CheY protein from <i>Thermotoga maritima</i> and <i> Escherichia coli</i>, and found no increase in ion pairs, ion-pair networks or hydrogen bonding.<sup>[http://onlinelibrary.wiley.com/doi/10.1002/pro.5560070221/abstract 20]</sup> Rather, they observed a shortening of the N and C termini of the thermostable protein, truncation of one of its loops and an increase in proline residues, all [http://en.wikipedia.org/wiki/Entropic_force entropic] factors. On the low end of the temperature scale, the inverse effect has been observed. From comparisons of structure from mesophilic or thermophilic <i>versus</i> psychrophilic organisms the psychrophilic proteins are found to have insertions in exposed loop regions.<sup>[http://www.pnas.org/content/89/11/5142 21]</sup> | ||
<br><br> | |||
Sequence alignments to proteins of known structure indicate that thermophilic sequences are more likely than their mesophilic homologs to have deletions in exposed loop regions. Loops were identified as the intervening regions between transmembrane segments.<sup>[http://www.sciencedirect.com/science/article/pii/S0022283603013147 3]</sup> This finding indicates a general evolutionary strategy for increasing thermostability and is thought to be a mechanism for reducing unfolded state entropy. By employing loop deletions as a mechanism for protein stability, an organisms should be able to withstand higher temperatures without protein denaturation<sup>[http://www.sciencedirect.com/science/article/pii/S0022283699928899 4]</sup><br> | Sequence alignments to proteins of known structure indicate that thermophilic sequences are more likely than their mesophilic homologs to have deletions in exposed loop regions. Loops were identified as the intervening regions between transmembrane segments.<sup>[http://www.sciencedirect.com/science/article/pii/S0022283603013147 3]</sup> This finding indicates a general evolutionary strategy for increasing thermostability and is thought to be a mechanism for reducing unfolded state entropy. By employing loop deletions as a mechanism for protein stability, an organisms should be able to withstand higher temperatures without protein denaturation<sup>[http://www.sciencedirect.com/science/article/pii/S0022283699928899 4]</sup> | ||
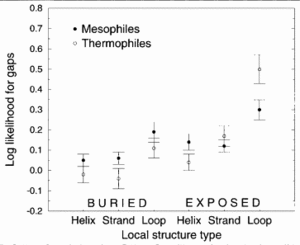
<br>Figure 4 demonstrates that thermophilic sequences have an increased propensity for deletions in exposed loops.<sup>[http://www.sciencedirect.com/science/article/pii/S0022283699928899 4]</sup> In the study conducted by Mandrich et al., three types of secondary structure, helix, strand, and loop, were considered. The figure depicts the structural propensities for gaps found in alignments between proteins of known structure and their mesophilic or thermophilic homologs. Propensities less than 0 for a given structure type indicate that fewer gaps are associate with that structure type than random expectation while higher propensities indicate that more gaps are associated. Here, the propensities were averaged over homologs for each protein of known structure. | <br><br> | ||
<br> | [[Image:loop deletions.gif|thumb|300px|left|Figure 4. Structural propensities for gaps found in alignments between proteins of known structure and their mesophilic or thermophilic homolog. Thompson and Eisenberg 1999.]] Figure 4 demonstrates that thermophilic sequences have an increased propensity for deletions in exposed loops.<sup>[http://www.sciencedirect.com/science/article/pii/S0022283699928899 4]</sup> In the study conducted by Mandrich <i>et al.</i>, three types of secondary structure, helix, strand, and loop, were considered. The figure depicts the structural propensities for gaps found in alignments between proteins of known structure and their mesophilic or thermophilic homologs. Propensities less than 0 for a given structure type indicate that fewer gaps are associate with that structure type than random expectation while higher propensities indicate that more gaps are associated. Here, the propensities were averaged over homologs for each protein of known structure. To add to the argument that thermophiles employ loop deletions for protein stabilization, the opposite effect is observed with organisms that live in cold temperatures. Cold-adapted organisms have been observed to have insertions in exposed loop regions in psychrophilic proteins.<sup>[http://www.jbc.org/content/269/26/17448.long 17]</sup> This figure suggests that exposed loops of thermophiles are the only structura elements having significantly greater gaps compared to mesophiles.<sup>[http://www.sciencedirect.com/science/article/pii/S0022283603013147 3]</sup> | ||
<br><br> | |||
As a result, deletion of the exposed loop residues decrease the unfolding entropy while having minimal impact on the enthalpy of unfolding and will cause protein stabilization. A full thermodynamic argument for this conclusion can be found in Thomson and Eisenberg 1999.<sup>[http://www.sciencedirect.com/science/article/pii/S0022283699928899 4]</sup> <br> | |||
<br> | <br> | ||
| Line 63: | Line 66: | ||
Since the GC pair is bound by three hydrogen bonds while the adenine-thymine (AT) pair is bound by two hydrogen bonds, it is expected that organisms growing at higher temperature would have a higher proportion of GC than AT pairs. Primary literature has conflicting data on this discussion. | Since the GC pair is bound by three hydrogen bonds while the adenine-thymine (AT) pair is bound by two hydrogen bonds, it is expected that organisms growing at higher temperature would have a higher proportion of GC than AT pairs. Primary literature has conflicting data on this discussion. | ||
<br><br> | |||
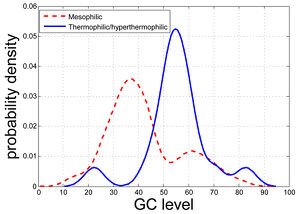
Hao and Wu found the GC content levels of the coding/non-coding regions of certain genes are highly likely to be correlated with the temperature range conditions of prokaryotic organisms. Four genes were consistently identified as correlated with the temperature range condition: K01251 (adenosylhomocysteinase), K03724 (DNA repair and recombination proteins), K07588 (LAO/AO transport system kinase), and K09122 (hypothetical protein). When these four genomic regions were used to predict the temperature range condition of an organism, the prediction accuracy was 84.52% for complete genomes, 84.09% for the in-progress genomes, and 82.70% for the metagenomes. Considering that these four genes only account for less than 1% of all the 413 genomic regions potentially correlated with the temperature range condition but can to a great extent retain the prediction accuracy, we may interpret these four genomic regions as the core of the temperature range-correlated. Additionally, research by Musto and Naya <i>et al.</i> support this stance by suggesting that, when specific families of prokaryotes (i.e. bacteria and archaea) are analyzed, there may be significant increases in GC content that coincide with an increase in optimal growth temperature. | [[Image:GC.jpg|thumb|300px|right|Figure 5. The distribution of the GC content level of the non-coding region surrounding the menB gene (K01661: naphthoate synthase) for mesophilic (dashed, red) and thermophilic/hyperthermophilic organisms (solid, blue). Hao and Wu 2010.]] | ||
Hao and Wu found the GC content levels of the coding/non-coding regions of certain genes are highly likely to be correlated with the temperature range conditions of prokaryotic organisms. These authors were inspired by preliminary studies, one of which observed that the non-coding region surrounding the gene menB (naphthoate synthase) can be drastically different for mesophilic and thermophilic/hyperthermophilic organisms (Figure 5).<sup>[http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3024870/ 22]</sup> Four genes were consistently identified as correlated with the temperature range condition: K01251 (adenosylhomocysteinase), K03724 (DNA repair and recombination proteins), K07588 (LAO/AO transport system kinase), and K09122 (hypothetical protein). When these four genomic regions were used to predict the temperature range condition of an organism, the prediction accuracy was 84.52% for complete genomes, 84.09% for the in-progress genomes, and 82.70% for the metagenomes. Considering that these four genes only account for less than 1% of all the 413 genomic regions potentially correlated with the temperature range condition but can to a great extent retain the prediction accuracy, we may interpret these four genomic regions as the core of the temperature range-correlated. Additionally, research by Musto and Naya <i>et al.</i> support this stance by suggesting that, when specific families of prokaryotes (i.e. bacteria and archaea) are analyzed, there may be significant increases in GC content that coincide with an increase in optimal growth temperature. | |||
<br><br> | |||
Hickey and Singer suggest that the large variations in the average genomic GC content between species are largely the result of bias mutation and repair pressures. Indeed, many highly thermophilic species, such as <i>Pyrococcus abyssi</i> and <i>Aquifex aeolicus</i>, have genomic GC contents of less than 50%, while some mesophiles - such as the human parasite <i>Mycobacterium tuberculosis</i> - have much higher GC contents in their genomes. This observation suggests that thermophiles have mechanisms other than increasing GC content for maintaining the double-stranded structure of their DNA at high temperatures.<sup>[http://www.ncbi.nlm.nih.gov/pmc/articles/PMC545586/ 7]</sup> | Hickey and Singer suggest that the large variations in the average genomic GC content between species are largely the result of bias mutation and repair pressures. Indeed, many highly thermophilic species, such as <i>Pyrococcus abyssi</i> and <i>Aquifex aeolicus</i>, have genomic GC contents of less than 50%, while some mesophiles - such as the human parasite <i>Mycobacterium tuberculosis</i> - have much higher GC contents in their genomes. This observation suggests that thermophiles have mechanisms other than increasing GC content for maintaining the double-stranded structure of their DNA at high temperatures.<sup>[http://www.ncbi.nlm.nih.gov/pmc/articles/PMC545586/ 7]</sup> | ||
| Line 73: | Line 77: | ||
==Conclusion== | ==Conclusion== | ||
The employment of structural salt bridges, loop deletions, polar charged residues, and heightened GC content have all been observed to heighten protein or DNA stability under high temperatures. The increased intramolecular forces, like ion and hydrogen bonds, of thermophilic structures are keystone to their thermostability. Astrobiologist can use these structural motifs to hypothesize and investigate possible extraterrestrial life forms. Perhaps planets that have a similar hot environment, like those of deep-sea hydrothermal vents or hot springs, can harbor organisms similar to the thermophiles we see on Earth. Although it is important to take into account that extraterrestrial life may not employ the same mechanisms of genetic material, metabolic processes, or protein structure, the study of thermophilic organisms opens a window into the amazing mechanisms employed in order to live in extreme environments. <br> | The employment of structural salt bridges, loop deletions, polar charged residues, disulfide bonding, and heightened GC content have all been observed to heighten protein or DNA stability under high temperatures. The increased intramolecular forces, like ion and hydrogen bonds, of thermophilic structures are keystone to their thermostability. Astrobiologist can use these structural motifs to hypothesize and investigate possible extraterrestrial life forms and early life on Earth. Perhaps planets that have a similar hot environment, like those of deep-sea hydrothermal vents or hot springs, can harbor organisms similar to the thermophiles we see on Earth. Although it is important to take into account that extraterrestrial life may not employ the same mechanisms of genetic material, metabolic processes, or protein structure, the study of thermophilic organisms opens a window into the amazing mechanisms employed in order to live in extreme environments. In addition to this, thermostable proteins can be utilized in the laboratory where many reactions need to be catalyzed with high temperatures. Using mesophilic proteins are not as effective in this sense because they are prone to denature at high temperatures. Thermophilic organisms present a repertoire of machinery that can retain their function at high temperatures, thereby offering novel tools to molecular biological research.<br> | ||
==References== | ==References== | ||
| Line 117: | Line 121: | ||
<sup>21</sup> [http://www.pnas.org/content/89/11/5142 Daggett, Valerie, and Michael Levitt. "A model of the molten globule state from molecular dynamics simulations." Proceedings of the National Academy of Sciences 89, no. 11 (1992): 5142-5146.] | <sup>21</sup> [http://www.pnas.org/content/89/11/5142 Daggett, Valerie, and Michael Levitt. "A model of the molten globule state from molecular dynamics simulations." Proceedings of the National Academy of Sciences 89, no. 11 (1992): 5142-5146.] | ||
<sup>22</sup> [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3024870/ Zheng, Hao, and Hongwei Wu. "Gene-centric association analysis for the correlation between the guanine-cytosine content levels and temperature range conditions of prokaryotic species." BMC bioinformatics 11.Suppl 11 (2010): S7.] | |||
Edited by (Paloma Medina), a student of [http://www.jsd.claremont.edu/faculty/profile.asp?FacultyID=254/ Nora Sullivan] in BIOL187S (Microbial Life) in [http://www.jsd.claremont.edu/ The Keck Science Department of the Claremont Colleges] Spring 2013. | Edited by (Paloma Medina), a student of [http://www.jsd.claremont.edu/faculty/profile.asp?FacultyID=254/ Nora Sullivan] in BIOL187S (Microbial Life) in [http://www.jsd.claremont.edu/ The Keck Science Department of the Claremont Colleges] Spring 2013. | ||
<!--Do not edit or remove this line-->[[Category:Pages edited by students of Nora Sullivan at the Claremont Colleges]] | <!--Do not edit or remove this line-->[[Category:Pages edited by students of Nora Sullivan at the Claremont Colleges]] | ||
Latest revision as of 01:00, 29 April 2013
A thermophile is an organism that thrives at relatively high temperatures, between 45 and 122°C. Thermophiles are found in a number of marine and terrestrial geothermally-heated habitats including shallow terrestrial hot springs, hydrothermal vent systems, sediment from volcanic islands, and deep sea hydrothermal vents. As prerequisite for their survival, thermophiles contain enzymes that can function at high temperatures. Investigating the DNA and protein stability of thermophiles is important because they can reveal what extraterrestrial life may look like and can give insights in biotechnology.
Introduction
The National Center for Biotechnology Inforation (NCBI) Microbial Genome Project Database uses five terms to categorize the temperature range an organism grows at, where cryophilic refers to –30° to –2°C, psychrophilic refers to –1° to +10°C, mesophilic refers to +11° to +45°C, thermophilic refers to +46° to 75°C, and hyperthermophilic refers to above +75°C.
Thermophiles are found in geothermally heated regions of the Earth like deep-sea hydrothermal vents and the hot springs of Yellowstone National Park (Figure 1). The investigation of thermophilic structure and chemistry poses very promising and intriguing contributions to the scientific community. For one, some of the enzymes used in molecular biology, like DNA polymerases, have derived from investigating heat-stable enzymes. Perhaps the quintessential example of a successful biotechnological application of thermozymes is the use of Taq polymerase, isolated from Thermus aquaticus 6, for PCR.
In addition, astrobiologists look to understand the structural and genomic correlates of hyperthermostability to give indication to what life may look like on planets hotter than ours. Also, astrobiologists, including researchers from NASA, suggest that hot springs all over the world provide some of the best "doorways into early Earth." 7 Many scientists believe that life might have begun roughly 4 billion years ago in high temperature environments and that the first organisms might therefore be thermophiles. Not only does this give insight into the origin of life on Earth, but opens up a new realm of possibilities for life elsewhere in the universe.7
Over the last 20 years, researchers have begun to discover key physical adaptations of proteins and DNA that allow thermophiles to remain functional and alive at high temperatures. First, increasing the number of salt bridges is a driving force for enhancement of the thermotolerance of proteins from hyperthermophilic microorganisms.2 Second, research suggests that the replacement of polar noncharged residues by charged ones constitutes a major stabilization mechanisms in the proteins of hyperthermophilic organisms.3 Third, disulfide bond abundance has been observed increases as a function of the maxiumum growth temperature of an organisms increases.14 Fourth, thermophilic protein sequences are more likely than their mesophilic homologs to have deletions in exposed loop regions. Lastly, there is evidence that the guanine-cytosine (GC) content levels of the coding/non-coding regions of certain genes are highly likely to be correlated with the temperature range conditions of prokaryotic organisms.
Protein Stabilization
High temperatures can often denature vital enzymes and proteins. Heating affects the secondary structure of proteins, causing changes in the shape of the molecule. Specifically, heat disrupts hydrogen bonds and non-polar hydrophobic interactions. This occurs because heat increases the kinetic energy and cause the molecules to vibrate so rapidly and violently that the bonds are disrupted. Changing a proteins shape can alter its natural function thereby disrupting vital processes like metabolism. In order to withstand hot temperatures, thermophilic proteins can exhibit higher core hydrophobicity9, greater numbers of ionic interactions10, increased packing density11, additional networks of hydrogen bonds12, decreased lengths of surface loops4, stabilization by heat stable chaperones13, and increase in disulfide bond formation14 and a general shortening of length15.
Salt Bridges

A salt bridge is a combination of two noncovalent interactions: hydrogen bonding and electrostatic interactions. The optimization of electrostatic interactions by increasing the number of salt bridges is a driving force for enhancement of the thermotolerance of proteins from hyperthermophilic microorganisms. 2 This trend is less evident in thermophilic organisms and absent from mesophile-derived proteins. Salt bridges often occur between groups distant in the protein sequence and form cross-links that stabilize tertiary structure. This interaction can increase the kinetic barrier towards thermal inactivation or thermal unfolding and, thus, prevents proteins from denaturing at high temperatures.
Table 1 shows the number of salt bridges in select thermo- and hyperthermophilic organisms. Ns indicates the number of salt bridges, Nr represents the number of salt bridges statistically expected for that protein structure, and Topt represents the temperature of optimal growth for the protein. Proteins from hyperthermophilic organisms are characterized by an increased number of ion pairs with respect to the statistical expectance and/or the number of ion-pairs in their mesophilic counterparts. This finding suggests that electrostatic interactions are a principal factor responsible for the elevation of the melting temperature of proteins from hyperthermophilic organisms.
The figure and table to the right show that charged residues in the enzyme from Aquifex aeolicus replace most of the polar residues in the Bacillus subtilis enzyme. Specifically, the number of ion pairs in the protein from Aquifex is increased by >90%. Furthermore, melting point assessments are often employed in thermostability studies in order to examine the effect of structural changes. In this case, the corresponding change in melting temperature when B. subtilis to A. aeolicus was about 27°C. 2
Molecular dynamics calculations on a prototypical ion-pair model system have suggested that a sizeable energy barrier exists for the solvation of a salt bridge and that the height of this barrier increases with temperature. Interestingly, a similar barrier is not seen with isosteric hydrophobic groups. Thus, the heightened kinetic barrier towards protein unfolding is one of the mechanisms thermophiles can employ to stabilize their proteins and prevent them from denaturing.
Polar Charged Residues
The charged polar amino acid content is a genomic signature that can give strong indications to an organisms growth environment. The advantage of polar amino acids comes from the increased stability of coulombic interactions with increasing temperatures. Therefore, in most structures of hyperthermophilic proteins, the existence of long chains of ion pairs provide cooperative stabilization. 3 This finding implies the existence of global structural features associated with hyperthermostability common to thermophilic Bacteria and Achaea as described below.
The replacement of polar noncharged residues by charged ones constitutes a major stabilization mechanism in the proteins of hyperthermophilic organisms. Residue changes allow the stabilization of proteins through ion bonds. A stronger intramolecular interaction between the protein and itself decreases the effect of an intermolecular force such as heat. In other words, more heat is required to denature the protein if there are stronger intramolecular interactions taking place. The proteome analysis in amino acid classes indicated that hyperthermophilicity is characterized by a sharp increase of charged residues, Lys and Glu, at the expense of polar noncharged residues, mainly Gln (Figure 3). Figure 3A is a plot of the sum of the percentages of charged amino acids (Lys, Arg, Asp, Glu; CHA, blue), polar noncharged amino acids (Asn, Gln, Ser, Thr; POL, green), and of the difference of the two values (CH-POL, red). The mesophiles and hyperthermophiles are identified by MESO and HYPER, respectively. Figure 3B is a plot of the percentages of the various amino acids in mesophiles (blue) and hyperthermophiles (red). Figure 3C is a plot of the sum of percentages of the various amino acid classes in mesophiles (blue) and hyperthermophiles (red). A threshold is observed around 10% for extremeophiles Ch-Po values, while a higher limit of 5% characterizes mesophiles. 1Cambillau and Claverie suggest from this data that the difference between charged and polar noncharged amino acids (Ch-Po) is the best indicator of an organism’s lifestyle.1
Disulfide Bonding
Structural disulfide bonds are a covalent tertiary interaction in extracellular and compartmentalized proteins, acting to stabilize a folded protein structure. The disulfide bond stabilizes the folded form of a protein in several ways. First, it holds two portions of the protein together, baising the protein towards the folded topology. Second, the disulfide bond may form the nucleus of a hydrophobic core of the folded protein, i.e., local hydrophobic residues may condense around the disulfide bond and onto each other through hydrophobic interactions. Third, the disulfide bond link two segments of the protein chain, the disulfide bond increases the effective local concentration of protein residues and lowers the effective local concentration of water molecules.16
The specific distribution of disulfide bonds observed across prokaryotic genomes suggests specialization in strategies used by organisms to stabilize their proteins. Trends in pairwise amino acid proximities were measure for proteins from 199 distinct prokaryotic genomes, and close cysteine-cysteine pairings were interpreted as likely specific disulfide bonds in these organisms.14 Thermophiles exhibited a pronounced bias in the spatial proximity of cysteine-cysteine residues, supporting a role for disulfide bonds in these organisms. The predicted disulfide abundance (expressed as a proximity score for cysteine-cysteine pairs) is shown in Figure 3 as a function of the maximum growth temperature of each organism. Here, a plot of log ratios of disulfide richness is identified in thermophiles, both archaeal and bacterial.
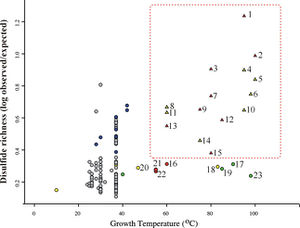
In addition to thermophilic prokaryotes, certain other organisms appear to have measurably elevated degrees of disulfide bonding. These include some halophiles, alkalophiles, acidophiles, and radiation-tolerant organisms. This trend suggests that disulfide bonds might serve generally to stabilize proteins in a variety of extreme environments.
Loop Deletions
There is evidence, both theoretical and experimental that deletion of exposed loop regions of protein structure can enhance stability. Simulations of protein unfolding have shown that unfoldings begin in exposed loop regions.18 Loop truncation has been a factor noted in several studies comparing crystal structures from mesophilic and thermophilic sources.19.
In one noteable example, Usher et al. compared the structures of the CheY protein from Thermotoga maritima and Escherichia coli, and found no increase in ion pairs, ion-pair networks or hydrogen bonding.20 Rather, they observed a shortening of the N and C termini of the thermostable protein, truncation of one of its loops and an increase in proline residues, all entropic factors. On the low end of the temperature scale, the inverse effect has been observed. From comparisons of structure from mesophilic or thermophilic versus psychrophilic organisms the psychrophilic proteins are found to have insertions in exposed loop regions.21
Sequence alignments to proteins of known structure indicate that thermophilic sequences are more likely than their mesophilic homologs to have deletions in exposed loop regions. Loops were identified as the intervening regions between transmembrane segments.3 This finding indicates a general evolutionary strategy for increasing thermostability and is thought to be a mechanism for reducing unfolded state entropy. By employing loop deletions as a mechanism for protein stability, an organisms should be able to withstand higher temperatures without protein denaturation4
Figure 4 demonstrates that thermophilic sequences have an increased propensity for deletions in exposed loops.4 In the study conducted by Mandrich et al., three types of secondary structure, helix, strand, and loop, were considered. The figure depicts the structural propensities for gaps found in alignments between proteins of known structure and their mesophilic or thermophilic homologs. Propensities less than 0 for a given structure type indicate that fewer gaps are associate with that structure type than random expectation while higher propensities indicate that more gaps are associated. Here, the propensities were averaged over homologs for each protein of known structure. To add to the argument that thermophiles employ loop deletions for protein stabilization, the opposite effect is observed with organisms that live in cold temperatures. Cold-adapted organisms have been observed to have insertions in exposed loop regions in psychrophilic proteins.17 This figure suggests that exposed loops of thermophiles are the only structura elements having significantly greater gaps compared to mesophiles.3
As a result, deletion of the exposed loop residues decrease the unfolding entropy while having minimal impact on the enthalpy of unfolding and will cause protein stabilization. A full thermodynamic argument for this conclusion can be found in Thomson and Eisenberg 1999.4
DNA Stabilization
In terms of DNA, the denaturation of nucleic acids is the separation of a double strand into two single strands, which occurs when the hydrogen bonds between the strands are broken. Denaturation of an organisms genetic material renders it unfit and highly susceptible to malicious mutations. Unlike most organisms, thermophiles can survive and thrive at very high temperatures.
GC Content
Since the GC pair is bound by three hydrogen bonds while the adenine-thymine (AT) pair is bound by two hydrogen bonds, it is expected that organisms growing at higher temperature would have a higher proportion of GC than AT pairs. Primary literature has conflicting data on this discussion.
Hao and Wu found the GC content levels of the coding/non-coding regions of certain genes are highly likely to be correlated with the temperature range conditions of prokaryotic organisms. These authors were inspired by preliminary studies, one of which observed that the non-coding region surrounding the gene menB (naphthoate synthase) can be drastically different for mesophilic and thermophilic/hyperthermophilic organisms (Figure 5).22 Four genes were consistently identified as correlated with the temperature range condition: K01251 (adenosylhomocysteinase), K03724 (DNA repair and recombination proteins), K07588 (LAO/AO transport system kinase), and K09122 (hypothetical protein). When these four genomic regions were used to predict the temperature range condition of an organism, the prediction accuracy was 84.52% for complete genomes, 84.09% for the in-progress genomes, and 82.70% for the metagenomes. Considering that these four genes only account for less than 1% of all the 413 genomic regions potentially correlated with the temperature range condition but can to a great extent retain the prediction accuracy, we may interpret these four genomic regions as the core of the temperature range-correlated. Additionally, research by Musto and Naya et al. support this stance by suggesting that, when specific families of prokaryotes (i.e. bacteria and archaea) are analyzed, there may be significant increases in GC content that coincide with an increase in optimal growth temperature.
Hickey and Singer suggest that the large variations in the average genomic GC content between species are largely the result of bias mutation and repair pressures. Indeed, many highly thermophilic species, such as Pyrococcus abyssi and Aquifex aeolicus, have genomic GC contents of less than 50%, while some mesophiles - such as the human parasite Mycobacterium tuberculosis - have much higher GC contents in their genomes. This observation suggests that thermophiles have mechanisms other than increasing GC content for maintaining the double-stranded structure of their DNA at high temperatures.7
Conclusion
The employment of structural salt bridges, loop deletions, polar charged residues, disulfide bonding, and heightened GC content have all been observed to heighten protein or DNA stability under high temperatures. The increased intramolecular forces, like ion and hydrogen bonds, of thermophilic structures are keystone to their thermostability. Astrobiologist can use these structural motifs to hypothesize and investigate possible extraterrestrial life forms and early life on Earth. Perhaps planets that have a similar hot environment, like those of deep-sea hydrothermal vents or hot springs, can harbor organisms similar to the thermophiles we see on Earth. Although it is important to take into account that extraterrestrial life may not employ the same mechanisms of genetic material, metabolic processes, or protein structure, the study of thermophilic organisms opens a window into the amazing mechanisms employed in order to live in extreme environments. In addition to this, thermostable proteins can be utilized in the laboratory where many reactions need to be catalyzed with high temperatures. Using mesophilic proteins are not as effective in this sense because they are prone to denature at high temperatures. Thermophilic organisms present a repertoire of machinery that can retain their function at high temperatures, thereby offering novel tools to molecular biological research.
References
16 "Disulfide Bond." Wikipedia. Wikimedia Foundation, 19 Apr. 2013. Web. 21 Apr. 2013.
Edited by (Paloma Medina), a student of Nora Sullivan in BIOL187S (Microbial Life) in The Keck Science Department of the Claremont Colleges Spring 2013.