Dengue virus envelope proteins: Difference between revisions
Cmazaher5634 (talk | contribs) No edit summary |
Cmazaher5634 (talk | contribs) |
||
| (46 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
{{Uncurated}} | {{Uncurated}} | ||
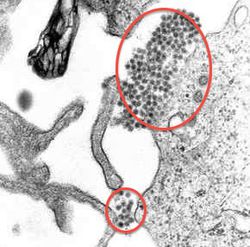
<br><b>Envelope proteins (E proteins)</b> are proteins on the surface on the dengue virus that allow it to interact with host cells. They are about 53 KDa and composed of three domains. In certain types of viruses, they are [http://en.wikipedia.org/wiki/Glycoprotein glycoproteins].([http://www.sciencedirect.com/science/article/pii/S1879625712000387 Pierson, 2012]) | <br><b>Envelope proteins (E proteins)</b> are proteins on the surface on the dengue virus that allow it to interact with host cells. They are about 53 KDa and composed of three domains. In certain types of viruses, they are [http://en.wikipedia.org/wiki/Glycoprotein glycoproteins]. ([http://www.sciencedirect.com/science/article/pii/S1879625712000387 Pierson, 2012]) A TEM image of dengue virions can be found in Figure 1. <br> | ||
[[Image:Dengue. | [[Image:Dengue annotated.jpeg|thumb|250px|right|<b>Figure 1.</b>TEM micrograph of dengue virus. The virions are present in the center of the image as dark dots. Red circles have been added to the original image to emphasize the locations of the virions. Originally by CDC per University of South Carolina Biomedical Sciences web page found through http://en.wikipedia.org/wiki/Dengue_fever ]] | ||
==<b>Background on Dengue Virus</b>== | ==<b>Background on Dengue Virus</b>== | ||
<br>Dengue is a term for a group of illnesses caused by four viruses, which are extremely relevant because they can infect humans. ([http://www.cdc.gov/dengue/ Dengue],[http://www.cdc.gov/dengue/epidemiology/index.html Epidemiology]) The | <br>Dengue is a term for a group of illnesses caused by four viruses, which are extremely relevant because they can infect humans. ([http://www.cdc.gov/dengue/ Dengue], [http://www.cdc.gov/dengue/epidemiology/index.html Epidemiology]) The dengue viruses belong to the [http://en.wikipedia.org/wiki/Flavivirus/ flavivirus] family, which are positive strand RNA viruses.([http://www.sciencedirect.com/science/article/pii/S1879625712000387 Pierson, 2012]) The viruses are known as DENV 1, DENV 2, DENV 3, and DENV 4. The primary vector of transmission is one of two types of [http://www.cdc.gov/dengue/resources/30Jan2012/comparisondenguevectors.pdf mosquitos], <i>Aedes aegypti</i> and <i>Aedes albopictus</i>. Although the viruses developed in humans between 100 and 800 years ago, dengue was not a prominent illness until recently. The emergence of dengue as an illness is thought to trace to increased travel during World War II leading the a wider distribution of the virus. In addition, lax efforts to control viral spread by eliminating the vectors has contributed to the incidence of dengue. ([http://www.cdc.gov/dengue/epidemiology/index.html Epidemiology], [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3317058/ Alen, 2012]) <br> | ||
<br>Dengue can manifest in two forms: dengue fever and dengue hemorrhagic fever (DHF). ([http://www.cdc.gov/Dengue/faqFacts/index.html Frequently]) | <br>Dengue can manifest in two forms: dengue fever and dengue hemorrhagic fever (DHF). ([http://www.cdc.gov/Dengue/faqFacts/index.html Frequently]) The [http://www.cdc.gov/Dengue/symptoms/index.html/ symptoms] of dengue infection include high fever, severe headache, eye pain, joint pain, muscle pain, bone pain, and rash. [http://www.cdc.gov/Dengue/faqFacts/index.html/ Education] about the illness and the risk of developing the more serious dengue hemorrhagic fever can benefit people who show signs of dengue infection. Even though few patients develop DHF, people who show symptoms should see a health professional for assessment in order to treat DHF early. DHF can have [http://www.nlm.nih.gov/medlineplus/ency/article/001373.htm/ additional symptoms] including restlessness with [http://en.wikipedia.org/wiki/Ecchymosis/ ecchymosis], rash, [http://en.wikipedia.org/wiki/Petechia/ petechiae], or shock with clammy extremities or sweating. Currently, there is no medicine to target either type of infection. In order to design a treatment for dengue infection, it is important to understand the mechanism of infection and the structure of the dengue virus family so that effective treatments can be designed. ([http://www.cdc.gov/Dengue/symptoms/index.html/ Symptoms], [http://www.nlm.nih.gov/medlineplus/ency/article/001373.htm/ Dengue hemorrhagic], and [http://www.cdc.gov/Dengue/faqFacts/index.html/ frequently]) <br> | ||
===<b>Role of E Proteins in Dengue Infection</b>=== | ===<b>Role of E Proteins in Dengue Infection</b>=== | ||
[[Image:CMV_schema.png|thumb|300px|right|Diagram of a cytomegalovirus | [[Image:CMV_schema.png|thumb|300px|right|<b>Figure 2.</b>Diagram of a cytomegalovirus. The image shows two types of glycoproteins, Glycoproteins I and III on the surface of the vision as well as other viral features, such as the capsid, genome, and coat. Source page: "Capsid." Wikipedia. http://en.wikipedia.org/wiki/Coat_protein ]] | ||
Envelope proteins are on the surface of the dengue virion and play a key role in cell entry; the structure of the protein will affect the way that the virus can interact with the host cell. | Envelope proteins are on the surface of the dengue virion and play a key role in cell entry; the structure of the protein will affect the way that the virus can interact with the host cell. Figure 2 shows a general schema of a virus with glycoproteins (such as E proteins) on the exterior of the cell. The dengue virus consists of the genome and nucleocapsid, which is the genetic material and surrounding proteins, contained within a viral envelope made of lipids. The viral envelope anchors multiple proteins including the E protein. ([http://www.nature.com/scitable/topicpage/dengue-viruses-22400925 Dengue Viruses])Being a flavivirus, dengue enters the host cell by means of receptor-mediated [http://en.wikipedia.org/wiki/Endocytosis endocytosis], meaning that the virus binds to receptor proteins on the cell to induce a conformational change that allows entry. ([http://www.pnas.org/content/100/12/6899.full/ Rey]) <br> | ||
<br>E proteins play a vital role in the dengue lifecycle by allowing the virus to enter host cells. As such, changes to the E protein may influence the course of the virus. The E proteins undergo conformational changes during endocytosis, and targeting the proteins may prove to be an effective antiviral strategy agains dengue. Although current research illuminates many new aspects about the role of E proteins, the specific mechanisms by which they function is still being elucidated.<br><br> | |||
| Line 19: | Line 20: | ||
==<b>Envelope Protein Variation</b>== | ==<b>Envelope Protein Variation</b>== | ||
<br>E protein structure varies between the four types of dengue and between individual strains of the same type. This variation manifests as changes in protein sequence due to genomic changes, as well as in three dimensional structural differences. <br> | <br>E protein structure varies between the four types of dengue and between individual strains of the same type. This variation manifests as changes in protein sequence due to genomic changes, as well as in three-dimensional structural differences. It is unclear the mechanism by which changing the envelope protein changes the pathogenicity of DENV strains, but strains with different degrees of pathogenicity often have differing E proteins.<br> | ||
===<b>Sequence Variation</b>=== | ===<b>Sequence Variation</b>=== | ||
<br>The sequence of the dengue virus envelope protein has been shown to vary strain to strain, but a direct link between sequence variation and pathogenesis has not been found. <br> | |||
<br>A study looked at | <br>A study looked at properties of the dengue virus, such as the E protein, that affect their degree of pathogenicity. Specifically, the authors looked at factors that determined whether the infection would manifest as dengue fever or dengue hemorrhagic fever. Since both immune system factors and viral factors determined the severity of symptoms, they focused their work on the viral factors. Using reverse transcriptase PCR, they compared the RNA genomes from different strains of DENV 2 obtained through patient plasma. Eleven strains were compared, some of which caused dengue fever and others which cause dengue hemorrhagic fever. The researchers noted that in most viruses related to dengue, “the molecular markers for pathogenicity have been localized in the E gene” meaning that if one strain is pathogenic and another is not there tends to be a difference in the part of the genome that codes for E proteins. ([http://jvi.asm.org/content/73/6/4738.short Leitmeyer, 1999])<br> | ||
<br>To further explore these molecular markers, the group sequenced the genomes and compared sequences to determine degree of similarity and mutation rates in samples from Southeast Asia and samples from the Americas. The Southeast Asian strains tended to be associated with dengue hemorrhagic fever while the American strains were not. The group showed that amino acids that related to the activity of the viruses were highly conserved, but that approximately fifty amino acid changes could be found. The phylogenetic tree produced from the results was consistent with previous predictions except that there was a high statistical significance for the separation of two genotypic groups. In the Southeast Asian group, there was no segregation based on resulting symptoms, and within the American group there was so segregation and all produced the same symptoms. The researchers were unable to find specific sequences that corresponded to type of symptom. This means that all viruses from certain groups, such as the Southeast Asian group, are capable of inducing dengue hemorrhagic fever. ([http://jvi.asm.org/content/73/6/4738.short Leitmeyer, 1999])<br> | <br>To further explore these molecular markers, the group sequenced the genomes and compared sequences to determine degree of similarity and mutation rates in samples from Southeast Asia and samples from the Americas. The Southeast Asian strains tended to be associated with dengue hemorrhagic fever while the American strains were not. The group showed that amino acids that related to the activity of the viruses were highly conserved, but that approximately fifty amino acid changes could be found. The phylogenetic tree produced from the results was consistent with previous predictions except that there was a high statistical significance for the separation of two genotypic groups. In the Southeast Asian group, there was no segregation based on resulting symptoms, and within the American group there was so segregation and all produced the same symptoms. The researchers were unable to find specific sequences that corresponded to type of symptom. This means that all viruses from certain groups, such as the Southeast Asian group, are capable of inducing dengue hemorrhagic fever. ([http://jvi.asm.org/content/73/6/4738.short Leitmeyer, 1999])<br> | ||
| Line 32: | Line 33: | ||
===<b>Structural Variation</b>=== | ===<b>Structural Variation</b>=== | ||
<br>Differing sequences of envelope proteins can affect their conformational arrangements, which in turn may alter their functionalities.<br> | |||
<br>The four different types of dengue virus have slightly different genomes, which means that they may have different protein structures because the genomic sequence for an E protein will alter the amino acid sequence and the folding of the protein. Antibodies that neutralize one type of dengue virus may be ineffective against another dengue strain due to these structural differences so understanding structural differences is key to treating the virus. Crystallography can show the folded structure of the proteins and allow for comparison of proteins from the different strains. Modis and collaborators used crystallography to learn about the structure of DENV 3 E protein and compare it to the previously characterized analogue in DENV 2. ([http://jvi.asm.org/content/79/2/1223.short Modis, 2005])<br> | <br>The four different types of dengue virus have slightly different genomes, which means that they may have different protein structures because the genomic sequence for an E protein will alter the amino acid sequence and the folding of the protein. Antibodies that neutralize one type of dengue virus may be ineffective against another dengue strain due to these structural differences so understanding structural differences is key to treating the virus. Crystallography can show the folded structure of the proteins and allow for comparison of proteins from the different strains. Modis and collaborators used crystallography to learn about the structure of DENV 3 E protein and compare it to the previously characterized analogue in DENV 2. ([http://jvi.asm.org/content/79/2/1223.short Modis, 2005])<br> | ||
<br>The researchers were able to crystallize a soluble DENV 3 E protein and determine the structure. Comparison of the E protein from DENV 3 and DENV 2 (strains CH53498 and PR159S1, respectively) lead to interesting insights. The two strains compared had 68% sequence agreement, and folded in similar ways. In solution, the proteins dimerized. ([http://jvi.asm.org/content/79/2/1223.short Modis, 2005])<br> | <br>The researchers were able to crystallize a soluble DENV 3 E protein and determine the structure. Comparison of the E protein from DENV 3 and DENV 2 (strains CH53498 and PR159S1, respectively) lead to interesting insights. The two strains compared had 68% sequence agreement, and folded in similar ways. In solution, the proteins dimerized. ([http://jvi.asm.org/content/79/2/1223.short Modis, 2005])<br> | ||
[[Image:Locations of asn residues.png|thumb|400px|left|<b>Figure 3.</b>Crystal structure of dimeric E proteins, showing both the Asn-153 and the Asn-67 position of each protein. The different colors of the structure represent different domains, or structural areas, of the proteins. Adapted from ([http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3317058/ Alen, 2012])]] | |||
<br>Modis and collaborators also explored the two <i>N</i>-linked glycosylation sites on the glycoproteins. The two resides where this occurs are Asn-67 and Asn-153. Asn-67 is unique to dengue viruses, while the second site can be found in a larger class of viruses. | <br>Modis and collaborators also explored the two <i>N</i>-linked glycosylation sites on the glycoproteins. The two resides where this occurs are Asn-67 and Asn-153. Asn-67 is unique to dengue viruses, while the second site can be found in a larger class of viruses. An image showing the locations of these residues on a dimer protein structure can be seen to the left in Figure 3. Researchers are interested in these glycosylation sites because the glycans on the protein helps the viruses successfully attack cells. In DENV 3, the protein loop with Asn-153 is shorter than in the other dengue viruses. Both Asn sites have glycans attached to them, which means that at least in <i>Drosophila</i>, the organism from which the protein was isolated, they are both necessary for infection. When DENV 2 was crystallized from<i> Aedes allobpicus</i>, there was no sugar at the 153 position. The group explained that the DENV 3 glycans may have crystallized well due to the other parts of the protein that they link to. The group notes that the most major difference between E proteins on the two types of dengue is the spatial orientation of domains I and II. The change in orientation between the two viruses is large enough that it must be attributed to a structural difference and not to protein movement in the original solutions. They note that within the domains, the high sequence agreement leads to some structural similarity. There are however, ([http://jvi.asm.org/content/79/2/1223.short Modis, 2005])<br> | ||
<br><br> | <br><br> | ||
| Line 42: | Line 45: | ||
==<b>Structural Changes of the Protein</b>== | ==<b>Structural Changes of the Protein</b>== | ||
Throughout the dengue lifecycle, the E proteins change conformations for a variety of reasons. During endocytosis, conformational changes of the E protein facilitate entry into the host cell. | Throughout the dengue lifecycle, the E proteins change conformations for a variety of reasons. During endocytosis, conformational changes of the E protein facilitate entry into the host cell. | ||
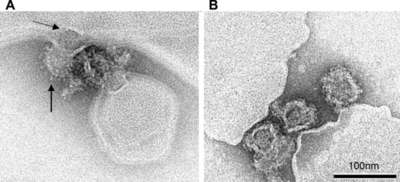
[[Image:micrograph dengue.png|thumb|400px|right|<b>Figure 4.</b>Micrograph of dengue virus fusing to liposome at a low pH. <b>a.</b>the lower arrow points to projections on the virion surface in response to acidity while the top arrow points to a structure that is probably the nucleocapsid. <b>b.</b>virions attached to a liposome in a basic solution. Adapted from ([http://www.plospathogens.org/article/info%3Adoi%2F10.1371%2Fjournal.ppat.0030020 Stiasny, 2007)] ]] <br> | |||
===<b>Fusion with Host Cell</b>=== | ===<b>Fusion with Host Cell</b>=== | ||
<br>E proteins have been implicated in the virus’s ability to bind to host cells, the mechanism for which is important for understanding the attack on the host. In order to better understand the mechanism, Klein and coworkers studied crystal structures | <br>E proteins have been implicated in the virus’s ability to bind to host cells, the mechanism for which is important for understanding the attack on the host. Figure 4 shows dengue virions entering the host. In order to better understand the binding mechanism, Klein and coworkers studied crystal structures to understand the final step of membrane fusion. The protein transitions from a dimeric form, shown in Figure 3, to a trimeric form in the process and the group was able to crystalize portions of this transition. ([http://jvi.asm.org/content/87/4/2287.short Klein, 2013])<br> | ||
==<b>E Proteins as Antiviral Targets</b>== | ==<b>E Proteins as Antiviral Targets</b>== | ||
<br>Given their key role in host cell entry, antiviral measures that target E proteins may be very effective. However, the variation between E proteins makes it difficult to use antiviral mechanisms, such as antibodies, to one strain of dengue against another strain. <br> | <br>Given their key role in host cell entry, antiviral measures that target E proteins may be very effective. However, the variation between E proteins makes it difficult to use antiviral mechanisms, such as antibodies, to one strain of dengue against another strain. <br> | ||
===<b>Immune Recognition</b>=== | ===<b>Immune Recognition</b>=== | ||
<br>An infected host may have an immune response to DENV centered around E protein interactions, with varying degrees of success at fighting the infection.<br> | |||
<br>The varied structures of E proteins have implications for host-virus interactions, as the body may create antibodies to try and counter dengue infection. Antibodies to the dengue viruses may have different affinities to different types of dengue due to structural features on the E proteins. Some of these features may be due variation of 56 amino acid residues in parts of the E proteins, especially variation in a portion of the protein that interacts with cell receptors. If antibodies cannot properly neutralize to the dengue virus, the presence of the antibody on the virus may enhance the virus’s ability to infiltrate the target cells because the host cells will recognize the antibodies as host material. This is called “antibody-dependent enhancement” of infection and can cause more severe symptoms. Other antibodies inhibit the viruses at regions that are conserved in the different types of dengue and tend work by inhibiting membrane fusion. ([http://jvi.asm.org/content/79/2/1223.short Modis, 2005])<br> | <br>The varied structures of E proteins have implications for host-virus interactions, as the body may create antibodies to try and counter dengue infection. Antibodies to the dengue viruses may have different affinities to different types of dengue due to structural features on the E proteins. Some of these features may be due variation of 56 amino acid residues in parts of the E proteins, especially variation in a portion of the protein that interacts with cell receptors. If antibodies cannot properly neutralize to the dengue virus, the presence of the antibody on the virus may enhance the virus’s ability to infiltrate the target cells because the host cells will recognize the antibodies as host material. This is called “antibody-dependent enhancement” of infection and can cause more severe symptoms. Other antibodies inhibit the viruses at regions that are conserved in the different types of dengue and tend work by inhibiting membrane fusion. ([http://jvi.asm.org/content/79/2/1223.short Modis, 2005])<br> | ||
<br> | <br> | ||
<br>Currently, there are multiple entry | ===<b>Current Antiviral Molecules</b>=== | ||
[[Image:Table 2 Alen 2012.png|thumb|400px|right| Portion of Table 2 from ([http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3317058/ Alen, 2012]) listing different types of antivirals for DENV. The first column lists type of antiviral. The second lists the specific compound. The third column states the type of DENV that the antiviral inhibits. The entire table can be found at ([http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3317058/ Alen, 2012]) ]] | |||
<br>Currently, there are multiple entry inhibitor molecules for dengue, but they have not been translated into effective antiviral treatments for patients. These molecules are early steps that may serve as the building blocks for future therapies. ([http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3317058/ Alen, 2012]) <br> | |||
[[Image:Table 2 Alen 2012.png|thumb|400px|right| <b>Figure 5.</b>Portion of Table 2 from ([http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3317058/ Alen, 2012]) listing different types of antivirals for DENV. The first column lists type of antiviral. The second lists the specific compound. The third column states the type of DENV that the antiviral inhibits. The entire table can be found at ([http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3317058/ Alen, 2012]) ]] | |||
<br>A group of the entry inhibitors acts mainly by interfering with structural changes to the E protein during cell entry. The entry inhibitors themselves do not serve as vaccines, but may be built on to further target dengue infection. In the | <br>A group of the entry inhibitors acts mainly by interfering with structural changes to the E protein during cell entry. The entry inhibitors themselves do not serve as vaccines, but may be built on to further target dengue infection. In the Figure 5, these types of inhibitors are classed as <i>Fusion inhibitors</i> ([http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3317058/ Alen, 2012]) <br> | ||
<br>Other antivirals inhibit proper formation of the viral proteins during replication in the host cell by stopping the <i>N</i>-linked glycosylation sites from forming on the new virus E proteins. Altering the protein formation could affect the ability of the virus to enter a new host cell. | <br>Other antivirals inhibit proper formation of the viral proteins during replication in the host cell by stopping the <i>N</i>-linked glycosylation sites from forming on the new virus E proteins. Altering the protein formation could affect the ability of the virus to enter a new host cell. Figure 5 classifies these drugs as <i>Glycosidase inhibitors</i>([http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3317058/ Alen, 2012]) <br> | ||
<br>Some antivirals also target the interaction between the sugars on the E proteins, the N-glycans, and the host cell. These therapies can be expensive, but are active against many strains of dengue. These antivirals are listed as <i>CBAs</i> in | <br>Some antivirals also target the interaction between the sugars on the E proteins, the N-glycans, and the host cell. These therapies can be expensive, but are active against many strains of dengue. These antivirals are listed as <i>CBAs</i> in Figure 5. ([http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3317058/ Alen, 2012])<br> | ||
<br>A final class of drugs acts by mimicking a host cell receptor, heparin sulfate (HS), and binding to the virus. While effective, these drugs tend to act as anticoagulants, which limits their usefulness in a clinical setting. Drugs that mimic heparin are called <i>Heparan mimetics</i> | <br>A final class of drugs acts by mimicking a host cell receptor, heparin sulfate (HS), and binding to the virus. While effective, these drugs tend to act as anticoagulants, which limits their usefulness in a clinical setting. Drugs that mimic heparin are called <i>Heparan mimetics</i> in Figure 5.([http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3317058/ Alen, 2012]) <br> | ||
<br><br> | <br><br> | ||
==<b>Conclusion</b>== | ==<b>Conclusion</b>== | ||
<br>Dengue is a group of viruses that infects humans via mosquito vectors. A structural feature of the virus that is essential for infection is the envelope protein, or E protein. While there are currently no drugs to specifically target the dengue viruses, substantial work is being done in order to understand the proteins that enable attack. Hopefully, these insights can lead to drug development | <br>Dengue is a group of viruses that infects humans via mosquito vectors. A structural feature of the virus that is essential for infection is the envelope protein, or E protein. While there are currently no drugs to specifically target the dengue viruses, substantial work is being done in order to understand the proteins that enable attack. Hopefully, these insights can lead to drug development by providing information on specific drug targets, however, the current research on the protein yields mostly comparative information between E proteins of different dengue strains and must be expanded. <br> | ||
==References== | ==References== | ||
| Line 98: | Line 103: | ||
===Photo credits=== | ===Photo credits=== | ||
<br><b>Figure | <br><b>Figure 1</b><br> | ||
<br> Source Page: "Dengue fever." Wikipedia. http://en.wikipedia.org/wiki/Dengue_fever <br> | |||
page used to retrieve image: File: Dengue.jpg Wikimedia Commons. http://upload.wikimedia.org/wikipedia/commons/b/b0/Dengue.jpg<br> | |||
<br><b>Figure 2: </b><br> | |||
<br>Source page: "Capsid." Wikipedia. http://en.wikipedia.org/wiki/Coat_protein<br> | <br>Source page: "Capsid." Wikipedia. http://en.wikipedia.org/wiki/Coat_protein<br> | ||
page used to retrieve image: File: CMVschema.svg. Wikipedia. http://upload.wikimedia.org/wikipedia/commons/thumb/9/91/CMVschema.svg/500px-CMVschema.svg.png<br><br> | page used to retrieve image: File: CMVschema.svg. Wikipedia. http://upload.wikimedia.org/wikipedia/commons/thumb/9/91/CMVschema.svg/500px-CMVschema.svg.png<br> | ||
<br><b> Figure | |||
<br><b>Figure 3: </b><br> | |||
<br>[http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3317058/ Alen, Marijke M. F. and Dominique Schols. (2012) Dengue Virus Entry as Target for Antivrial Therapy. Journal of Tropical Medicine. pages not listed.]<br> | |||
<br><b> Figure 4:</b><br> | |||
<br>[http://www.plospathogens.org/article/info%3Adoi%2F10.1371%2Fjournal.ppat.0030020 Stiasny K, Kössl C, Lepault J, Rey FA, Heinz FX (2007) Characterization of a Structural Intermediate of Flavivirus Membrane Fusion. PLoS Pathog 3(2): e20. doi:10.1371/journal.ppat.0030020]<br> | |||
<br><b> Figure 5:</b><br> | |||
<br>[http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3317058/ Alen, Marijke M. F. and Dominique Schols. (2012) Dengue Virus Entry as Target for Antivrial Therapy. Journal of Tropical Medicine. pages not listed.]<br> | <br>[http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3317058/ Alen, Marijke M. F. and Dominique Schols. (2012) Dengue Virus Entry as Target for Antivrial Therapy. Journal of Tropical Medicine. pages not listed.]<br> | ||
Latest revision as of 06:58, 29 April 2013
Envelope proteins (E proteins) are proteins on the surface on the dengue virus that allow it to interact with host cells. They are about 53 KDa and composed of three domains. In certain types of viruses, they are glycoproteins. (Pierson, 2012) A TEM image of dengue virions can be found in Figure 1.
Background on Dengue Virus
Dengue is a term for a group of illnesses caused by four viruses, which are extremely relevant because they can infect humans. (Dengue, Epidemiology) The dengue viruses belong to the flavivirus family, which are positive strand RNA viruses.(Pierson, 2012) The viruses are known as DENV 1, DENV 2, DENV 3, and DENV 4. The primary vector of transmission is one of two types of mosquitos, Aedes aegypti and Aedes albopictus. Although the viruses developed in humans between 100 and 800 years ago, dengue was not a prominent illness until recently. The emergence of dengue as an illness is thought to trace to increased travel during World War II leading the a wider distribution of the virus. In addition, lax efforts to control viral spread by eliminating the vectors has contributed to the incidence of dengue. (Epidemiology, Alen, 2012)
Dengue can manifest in two forms: dengue fever and dengue hemorrhagic fever (DHF). (Frequently) The symptoms of dengue infection include high fever, severe headache, eye pain, joint pain, muscle pain, bone pain, and rash. Education about the illness and the risk of developing the more serious dengue hemorrhagic fever can benefit people who show signs of dengue infection. Even though few patients develop DHF, people who show symptoms should see a health professional for assessment in order to treat DHF early. DHF can have additional symptoms including restlessness with ecchymosis, rash, petechiae, or shock with clammy extremities or sweating. Currently, there is no medicine to target either type of infection. In order to design a treatment for dengue infection, it is important to understand the mechanism of infection and the structure of the dengue virus family so that effective treatments can be designed. (Symptoms, Dengue hemorrhagic, and frequently)
Role of E Proteins in Dengue Infection

Envelope proteins are on the surface of the dengue virion and play a key role in cell entry; the structure of the protein will affect the way that the virus can interact with the host cell. Figure 2 shows a general schema of a virus with glycoproteins (such as E proteins) on the exterior of the cell. The dengue virus consists of the genome and nucleocapsid, which is the genetic material and surrounding proteins, contained within a viral envelope made of lipids. The viral envelope anchors multiple proteins including the E protein. (Dengue Viruses)Being a flavivirus, dengue enters the host cell by means of receptor-mediated endocytosis, meaning that the virus binds to receptor proteins on the cell to induce a conformational change that allows entry. (Rey)
E proteins play a vital role in the dengue lifecycle by allowing the virus to enter host cells. As such, changes to the E protein may influence the course of the virus. The E proteins undergo conformational changes during endocytosis, and targeting the proteins may prove to be an effective antiviral strategy agains dengue. Although current research illuminates many new aspects about the role of E proteins, the specific mechanisms by which they function is still being elucidated.
Envelope Protein Variation
E protein structure varies between the four types of dengue and between individual strains of the same type. This variation manifests as changes in protein sequence due to genomic changes, as well as in three-dimensional structural differences. It is unclear the mechanism by which changing the envelope protein changes the pathogenicity of DENV strains, but strains with different degrees of pathogenicity often have differing E proteins.
Sequence Variation
The sequence of the dengue virus envelope protein has been shown to vary strain to strain, but a direct link between sequence variation and pathogenesis has not been found.
A study looked at properties of the dengue virus, such as the E protein, that affect their degree of pathogenicity. Specifically, the authors looked at factors that determined whether the infection would manifest as dengue fever or dengue hemorrhagic fever. Since both immune system factors and viral factors determined the severity of symptoms, they focused their work on the viral factors. Using reverse transcriptase PCR, they compared the RNA genomes from different strains of DENV 2 obtained through patient plasma. Eleven strains were compared, some of which caused dengue fever and others which cause dengue hemorrhagic fever. The researchers noted that in most viruses related to dengue, “the molecular markers for pathogenicity have been localized in the E gene” meaning that if one strain is pathogenic and another is not there tends to be a difference in the part of the genome that codes for E proteins. (Leitmeyer, 1999)
To further explore these molecular markers, the group sequenced the genomes and compared sequences to determine degree of similarity and mutation rates in samples from Southeast Asia and samples from the Americas. The Southeast Asian strains tended to be associated with dengue hemorrhagic fever while the American strains were not. The group showed that amino acids that related to the activity of the viruses were highly conserved, but that approximately fifty amino acid changes could be found. The phylogenetic tree produced from the results was consistent with previous predictions except that there was a high statistical significance for the separation of two genotypic groups. In the Southeast Asian group, there was no segregation based on resulting symptoms, and within the American group there was so segregation and all produced the same symptoms. The researchers were unable to find specific sequences that corresponded to type of symptom. This means that all viruses from certain groups, such as the Southeast Asian group, are capable of inducing dengue hemorrhagic fever. (Leitmeyer, 1999)
Within the two sample groups, there was an amino acid difference at the 390 position of the E protein; mutations in the protein had previously been shown to alter the virulence of dengue and viruses of the same family. The mutation occurred in a portion of the protein that tends to be folded towards the outer surface and that has been shown to interact with the host cell surface. In the American variety, which did not produce hemorrhagic fever, the 370 position was an Asp, while in the Southeast Asian variety, which could produce hemorrhagic fever, the 730 position was an Asn and is believed to interact more strongly with host cells. The researchers were able to identify differences in the sequence of the proteins, but the role of these differences in pathogenicity is not known. (Leitmeyer, 1999).
Structural Variation
Differing sequences of envelope proteins can affect their conformational arrangements, which in turn may alter their functionalities.
The four different types of dengue virus have slightly different genomes, which means that they may have different protein structures because the genomic sequence for an E protein will alter the amino acid sequence and the folding of the protein. Antibodies that neutralize one type of dengue virus may be ineffective against another dengue strain due to these structural differences so understanding structural differences is key to treating the virus. Crystallography can show the folded structure of the proteins and allow for comparison of proteins from the different strains. Modis and collaborators used crystallography to learn about the structure of DENV 3 E protein and compare it to the previously characterized analogue in DENV 2. (Modis, 2005)
The researchers were able to crystallize a soluble DENV 3 E protein and determine the structure. Comparison of the E protein from DENV 3 and DENV 2 (strains CH53498 and PR159S1, respectively) lead to interesting insights. The two strains compared had 68% sequence agreement, and folded in similar ways. In solution, the proteins dimerized. (Modis, 2005)
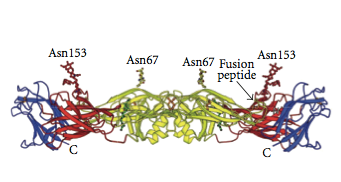
Modis and collaborators also explored the two N-linked glycosylation sites on the glycoproteins. The two resides where this occurs are Asn-67 and Asn-153. Asn-67 is unique to dengue viruses, while the second site can be found in a larger class of viruses. An image showing the locations of these residues on a dimer protein structure can be seen to the left in Figure 3. Researchers are interested in these glycosylation sites because the glycans on the protein helps the viruses successfully attack cells. In DENV 3, the protein loop with Asn-153 is shorter than in the other dengue viruses. Both Asn sites have glycans attached to them, which means that at least in Drosophila, the organism from which the protein was isolated, they are both necessary for infection. When DENV 2 was crystallized from Aedes allobpicus, there was no sugar at the 153 position. The group explained that the DENV 3 glycans may have crystallized well due to the other parts of the protein that they link to. The group notes that the most major difference between E proteins on the two types of dengue is the spatial orientation of domains I and II. The change in orientation between the two viruses is large enough that it must be attributed to a structural difference and not to protein movement in the original solutions. They note that within the domains, the high sequence agreement leads to some structural similarity. There are however, (Modis, 2005)
Structural Changes of the Protein
Throughout the dengue lifecycle, the E proteins change conformations for a variety of reasons. During endocytosis, conformational changes of the E protein facilitate entry into the host cell.
Fusion with Host Cell
E proteins have been implicated in the virus’s ability to bind to host cells, the mechanism for which is important for understanding the attack on the host. Figure 4 shows dengue virions entering the host. In order to better understand the binding mechanism, Klein and coworkers studied crystal structures to understand the final step of membrane fusion. The protein transitions from a dimeric form, shown in Figure 3, to a trimeric form in the process and the group was able to crystalize portions of this transition. (Klein, 2013)
E Proteins as Antiviral Targets
Given their key role in host cell entry, antiviral measures that target E proteins may be very effective. However, the variation between E proteins makes it difficult to use antiviral mechanisms, such as antibodies, to one strain of dengue against another strain.
Immune Recognition
An infected host may have an immune response to DENV centered around E protein interactions, with varying degrees of success at fighting the infection.
The varied structures of E proteins have implications for host-virus interactions, as the body may create antibodies to try and counter dengue infection. Antibodies to the dengue viruses may have different affinities to different types of dengue due to structural features on the E proteins. Some of these features may be due variation of 56 amino acid residues in parts of the E proteins, especially variation in a portion of the protein that interacts with cell receptors. If antibodies cannot properly neutralize to the dengue virus, the presence of the antibody on the virus may enhance the virus’s ability to infiltrate the target cells because the host cells will recognize the antibodies as host material. This is called “antibody-dependent enhancement” of infection and can cause more severe symptoms. Other antibodies inhibit the viruses at regions that are conserved in the different types of dengue and tend work by inhibiting membrane fusion. (Modis, 2005)
Current Antiviral Molecules
Currently, there are multiple entry inhibitor molecules for dengue, but they have not been translated into effective antiviral treatments for patients. These molecules are early steps that may serve as the building blocks for future therapies. (Alen, 2012)
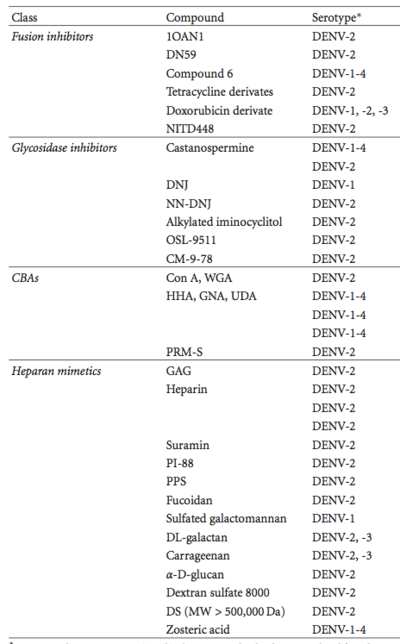
A group of the entry inhibitors acts mainly by interfering with structural changes to the E protein during cell entry. The entry inhibitors themselves do not serve as vaccines, but may be built on to further target dengue infection. In the Figure 5, these types of inhibitors are classed as Fusion inhibitors (Alen, 2012)
Other antivirals inhibit proper formation of the viral proteins during replication in the host cell by stopping the N-linked glycosylation sites from forming on the new virus E proteins. Altering the protein formation could affect the ability of the virus to enter a new host cell. Figure 5 classifies these drugs as Glycosidase inhibitors(Alen, 2012)
Some antivirals also target the interaction between the sugars on the E proteins, the N-glycans, and the host cell. These therapies can be expensive, but are active against many strains of dengue. These antivirals are listed as CBAs in Figure 5. (Alen, 2012)
A final class of drugs acts by mimicking a host cell receptor, heparin sulfate (HS), and binding to the virus. While effective, these drugs tend to act as anticoagulants, which limits their usefulness in a clinical setting. Drugs that mimic heparin are called Heparan mimetics in Figure 5.(Alen, 2012)
Conclusion
Dengue is a group of viruses that infects humans via mosquito vectors. A structural feature of the virus that is essential for infection is the envelope protein, or E protein. While there are currently no drugs to specifically target the dengue viruses, substantial work is being done in order to understand the proteins that enable attack. Hopefully, these insights can lead to drug development by providing information on specific drug targets, however, the current research on the protein yields mostly comparative information between E proteins of different dengue strains and must be expanded.
References
Cited in text
Alen, Marijke M. F. and Dominique Schols. (2012) Dengue Virus Entry as Target for Antivrial Therapy. Journal of Tropical Medicine. pages not listed.
"Dengue." Center for Disease Control and Prevention. 6 February 2013. http://www.cdc.gov/Dengue/
"Dengue hemorrhagic fever." MedlinePlus Medical Encyclopedia. U. S. National Library of Medicine. 22 March 2013. http://www.nlm.nih.gov/medlineplus/ency/article/001373.htm
"Dengue Viruses." Scitable. Nature Education. 2013. http://www.nature.com/scitable/topicpage/dengue-viruses-22400925
"Epidemiology." Dengue. Center for Disease Control and Prevention. 27 September 2012. http://www.cdc.gov/dengue/epidemiology/index.html
"Frequently Asked Questions." Dengue. Center for Disease Control and Prevention. 27 September 2012. http://www.cdc.gov/Dengue/faqFacts/index.html
Rey, Felix A. (2003) Dengue virus envelope glycoprotein structure: new insight into its interactions during viral entry. PNAS 100 (12) 6899-6901. Published online June 2, 2003. Published in print June 10, 2003. http://www.pnas.org/content/100/12/6899.full
Symptoms & Treatment. Center for Disease control and Prevention. http://www.cdc.gov/dengue/symptoms/
Photo credits
Figure 1
Source Page: "Dengue fever." Wikipedia. http://en.wikipedia.org/wiki/Dengue_fever
page used to retrieve image: File: Dengue.jpg Wikimedia Commons. http://upload.wikimedia.org/wikipedia/commons/b/b0/Dengue.jpg
Figure 2:
Source page: "Capsid." Wikipedia. http://en.wikipedia.org/wiki/Coat_protein
page used to retrieve image: File: CMVschema.svg. Wikipedia. http://upload.wikimedia.org/wikipedia/commons/thumb/9/91/CMVschema.svg/500px-CMVschema.svg.png
Link Credits
in order of appearance
"Glycoprotein." Wikipedia. http://en.wikipedia.org/wiki/Glycoprotein
"Comparison Between Main Dengue Vectors. Entomology." Dengue. Center for Disease Control and Prevention. http://www.cdc.gov/dengue/resources/30Jan2012/comparisondenguevectors.pdf
"Symptoms & Treatment." Dengue. Center for Disease Control and Prevention. 27 September 2012. http://www.cdc.gov/Dengue/symptoms/index.html/
"Frequently Asked Questions." Dengue. Center for Disease Control and Prevention. 27 September 2012. http://www.cdc.gov/Dengue/faqFacts/index.html
"Dengue hemorrhagic fever." MedlinePlus Medical Encyclopedia. U. S. National Library of Medicine. 22 March 2013. http://www.nlm.nih.gov/medlineplus/ency/article/001373.htm
"Ecchymosis." Wikipedia. http://en.wikipedia.org/wiki/Ecchymosis
"Petechia." Wikipedia. http://en.wikipedia.org/wiki/Petechia
"Endocytosis." Wikipedia. http://en.wikipedia.org/wiki/Endocytosis
Edited by Claire M, a student of Nora Sullivan in BIOL187S (Microbial Life) in The Keck Science Department of the Claremont Colleges Spring 2013.
