Spiroplasma melliferum: Difference between revisions
| (24 intermediate revisions by the same user not shown) | |||
| Line 5: | Line 5: | ||
==Classification== | ==Classification== | ||
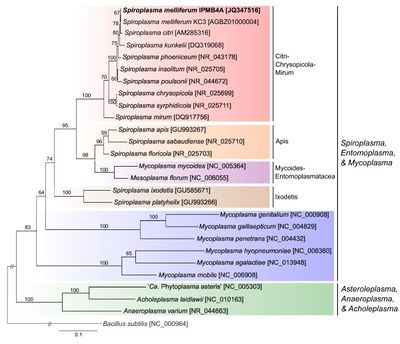
[[File:Phylogenetic_Placement.jpeg|thumb|right|400px|The Phylogenetic Placement of <I>Spiroplasma melliferum</I>]] | |||
===Higher Order Taxa:=== | ===Higher Order Taxa:=== | ||
| Line 19: | Line 17: | ||
Species: <I>melliferum </I> | Species: <I>melliferum </I> | ||
==Description and Significance== | ==Description and Significance== | ||
<I>Spiroplasma melliferum</I> is a bacterium that was isolated from hemolymph and gut of honeybees <I>(Apis mellifera)</I> as Strain BC-3<sup>T</sup> (= ATCC 33219). It was also recovered from hemolymph of bumble bees, leafcutter bees, and a robber fly and the intestinal contents of sweat bees, digger bees, bumblebees, and a butterfly. It was also found in a variety of plant surfaces (flowers) (2). | |||
<I>Spiroplasma melliferum</I> is susceptible to aminoglycoside antibiotics that contain a deoxystreptamine moiety such as: kanamycin, neomycin, and gentamicin whereas those that lack this moiety (e.g., kasugamycin, hygromycin and spectinomycin) are not effective (2). The presence of penicillin had no influence on growth rates. | |||
NCBI Accession #: JQ347516 | |||
==Genome Structure== | ==Genome Structure== | ||
The whole-genome shotgun sequencing of <I>S. Melliferum</I> IPMB4A produced a draft assembly that was ~1.1 Mb in size and covered ~80% of the chromosome (1). | |||
The G+C content of the DNA is 26 mol% as determined by the melting temperature method and 27 to 28 mol% as determined by the buoyant density method. The genome molecular weight is 10<sup>9</sup> (2). | |||
==Cell and Colony Structure== | |||
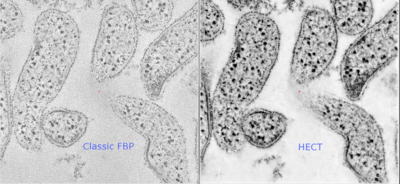
[[File:Spiroplasma_melliferum.png|thumb|right|400px|The Structure of the Cytoskeleton of <I>Spiroplasma melliferum</I> with High Efficiency CT (HECT).]] | |||
<I>Spiroplasma melliferum</I> cells are pleomorphic, varying from helical filaments that are 100 to 150 nm in diameter and 3 to 10 μm long to non-helical filaments or spherical cells that are 300 to 800 nm in diameter. They frequently have a fried-egg morphology. <I>Spiroplasma melliferum</I> are motile bacteria that lack true cell walls and periplasmic fibrils (2). | |||
<I>Spiroplasma melliferum</I> is a facultative anaerobe that is best grown in between 32 to 35°. Oleic acid, cholesterol, and bovine serum albumin were also essential for growth to occur (2). | |||
==Metabolism== | |||
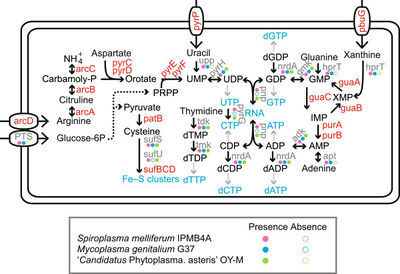
<I>Spiroplasma melliferum</I> | [[File:Metabolic_Pathways.jpeg|thumb|right|400px|The Metabolic Pathways of <I>Spiroplasma melliferum</I>]] | ||
<I>Spiroplasma melliferum</I> ferments glucose and hydrolyzes arginine (1). Acid is produced from glucose and mannose and does not hydrolyze urea (2). It also possesses all the genes required for glycolysis to convert glucose-6-phophate into pyruvate which is used for the production of cysteine (1). | |||
<I>Spiroplasma melliferum</I> cells extrude sodium ions by an energy dependent process and that the energy requirement for this process could be fulfilled by either glucose or arginine (3). | |||
==Ecology== | |||
Endosymbiotic inhabitants. | |||
== | ==Pathology== | ||
While most of the <I>Spiroplasma</I> species appeared to be harmless commensals of insects, a small number of species have evolved pathogenicity toward various arthropods and plants (1). | |||
This strain and other strains of the cluster entered the hemocoel of their insect hosts after per os acquisition, caused pathology in various tissues, and reduced adult longevity (2). | |||
== | ==References== | ||
1) Wen-Sui Lo, Ling-Ling Chen, Wan-Chia Chung, Gail Gasparich, Chih-Horng Kuo. (2013) <B>Comparative genome analysis of <I>Spiroplasma melliferum</I> IPMB4A, a honeybee-associated bacterium.</B> BMC Genomics. 9 January 2014. http://www.biomedcentral.com/1471-2164/14/22 | |||
2) Clark, T. B., R. F. Whitcomb, J. G. Tully, C. Mouches, C. Saillard, J. M. Bove, H. Wroblewski, P. Carle, D. L. Rose, R.B. Henegar, and D. L. Williamson. (1985) <B><I>Spiroplasma melliferum</I>, a New Species from the Honeybee <I>(Apis mellifera)</I>. </B>International Journal of Systematic Bacteriology, Vol. 35, No. 3, 296-308. | |||
3) Shirazi, Idit., Mark Tarshis and Shlomo Rottem. (1995) <B>An Arginine/ornithine Exchange System in <I>Spiroplasma melliferum</I>.</B> Microbiology, Vol. 141, 2323-2328. http://mic.sgmjournals.org/content/141/9/2323.full.pdf | |||
Edited by Brittany Demmons, student of Dr. Lisa R. Moore, University of Southern Maine, Department of Biological Sciences, http://www.usm.maine.edu/bio | |||
Latest revision as of 23:11, 12 January 2014
A Microbial Biorealm page on Spiroplasma melliferum
Classification
Higher Order Taxa:
Class: Mollicutes
Order: Mycoplasmatales
Family: Spiroplasmataceae
Genus: Spiroplasma
Species: melliferum
Description and Significance
Spiroplasma melliferum is a bacterium that was isolated from hemolymph and gut of honeybees (Apis mellifera) as Strain BC-3T (= ATCC 33219). It was also recovered from hemolymph of bumble bees, leafcutter bees, and a robber fly and the intestinal contents of sweat bees, digger bees, bumblebees, and a butterfly. It was also found in a variety of plant surfaces (flowers) (2).
Spiroplasma melliferum is susceptible to aminoglycoside antibiotics that contain a deoxystreptamine moiety such as: kanamycin, neomycin, and gentamicin whereas those that lack this moiety (e.g., kasugamycin, hygromycin and spectinomycin) are not effective (2). The presence of penicillin had no influence on growth rates.
NCBI Accession #: JQ347516
Genome Structure
The whole-genome shotgun sequencing of S. Melliferum IPMB4A produced a draft assembly that was ~1.1 Mb in size and covered ~80% of the chromosome (1).
The G+C content of the DNA is 26 mol% as determined by the melting temperature method and 27 to 28 mol% as determined by the buoyant density method. The genome molecular weight is 109 (2).
Cell and Colony Structure
Spiroplasma melliferum cells are pleomorphic, varying from helical filaments that are 100 to 150 nm in diameter and 3 to 10 μm long to non-helical filaments or spherical cells that are 300 to 800 nm in diameter. They frequently have a fried-egg morphology. Spiroplasma melliferum are motile bacteria that lack true cell walls and periplasmic fibrils (2).
Spiroplasma melliferum is a facultative anaerobe that is best grown in between 32 to 35°. Oleic acid, cholesterol, and bovine serum albumin were also essential for growth to occur (2).
Metabolism
Spiroplasma melliferum ferments glucose and hydrolyzes arginine (1). Acid is produced from glucose and mannose and does not hydrolyze urea (2). It also possesses all the genes required for glycolysis to convert glucose-6-phophate into pyruvate which is used for the production of cysteine (1).
Spiroplasma melliferum cells extrude sodium ions by an energy dependent process and that the energy requirement for this process could be fulfilled by either glucose or arginine (3).
Ecology
Endosymbiotic inhabitants.
Pathology
While most of the Spiroplasma species appeared to be harmless commensals of insects, a small number of species have evolved pathogenicity toward various arthropods and plants (1).
This strain and other strains of the cluster entered the hemocoel of their insect hosts after per os acquisition, caused pathology in various tissues, and reduced adult longevity (2).
References
1) Wen-Sui Lo, Ling-Ling Chen, Wan-Chia Chung, Gail Gasparich, Chih-Horng Kuo. (2013) Comparative genome analysis of Spiroplasma melliferum IPMB4A, a honeybee-associated bacterium. BMC Genomics. 9 January 2014. http://www.biomedcentral.com/1471-2164/14/22
2) Clark, T. B., R. F. Whitcomb, J. G. Tully, C. Mouches, C. Saillard, J. M. Bove, H. Wroblewski, P. Carle, D. L. Rose, R.B. Henegar, and D. L. Williamson. (1985) Spiroplasma melliferum, a New Species from the Honeybee (Apis mellifera). International Journal of Systematic Bacteriology, Vol. 35, No. 3, 296-308.
3) Shirazi, Idit., Mark Tarshis and Shlomo Rottem. (1995) An Arginine/ornithine Exchange System in Spiroplasma melliferum. Microbiology, Vol. 141, 2323-2328. http://mic.sgmjournals.org/content/141/9/2323.full.pdf
Edited by Brittany Demmons, student of Dr. Lisa R. Moore, University of Southern Maine, Department of Biological Sciences, http://www.usm.maine.edu/bio