Dehalobacter: Difference between revisions
| (15 intermediate revisions by the same user not shown) | |||
| Line 27: | Line 27: | ||
==Genome Structure== | ==Genome Structure== | ||
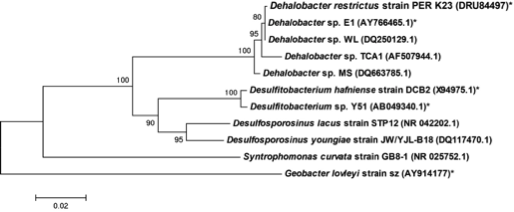
[[Image:16S_rRNA_based_classification_of_Dehalobacter_restrictus.png|frame| | [[Image:16S_rRNA_based_classification_of_Dehalobacter_restrictus.png|frame|below|16S rRNA based phylogenetic classification of Dehalobacter spp. From [Source, Kruse, T. et al., Standards in Genomic Sciences, 2013.]]] | ||
The whole genome of Dehalobacter restrictus PER- K23 was recently sequenced revealing critical insights into phylogeny and metabolic capabilities of the genus. The 2,943 kbp long genome contains 2,826 protein coding and 82 RNA genes, including 5 16S rRNA genes. Interestingly, the genome contains 25 predicted rdase genes, the majority of which appear to be full length. The rdase genes are mainly located in two clusters, suggesting a much larger potential for organohalide respiration than previously anticipated (11). | The whole genome of Dehalobacter restrictus PER- K23 was recently sequenced revealing critical insights into phylogeny and metabolic capabilities of the genus. The 2,943 kbp long genome contains 2,826 protein coding and 82 RNA genes, including 5 16S rRNA genes. Interestingly, the genome contains 25 predicted rdase genes, the majority of which appear to be full length. The rdase genes are mainly located in two clusters, suggesting a much larger potential for organohalide respiration than previously anticipated (11). | ||
==Cell Structure | ==Cell Structure== | ||
[[Image:close_up.GIF|frame|left|Electron micrograph of a thin-section of strain PER-K23, showing the cytoplasmic membrane (cm), the peptidoglycan layer (p), and the surface layer (s). From [http://lbewww.epfl.ch/LBE/PDF/Publi/CH5.pdf Holliger ''et al''.]]] | [[Image:close_up.GIF|frame|left|Electron micrograph of a thin-section of strain PER-K23, showing the cytoplasmic membrane (cm), the peptidoglycan layer (p), and the surface layer (s). From [http://lbewww.epfl.ch/LBE/PDF/Publi/CH5.pdf Holliger ''et al''.]]] Dehalobacter restrictus cells are rod shaped, 2-3 µm in length and 0.3-0.5 µm in diameter with tapering ends. There is a single lateral flagellum at one end indicating that the cells are motile. The cells do not form spores under substrate and temperature induced stress. Cell wall stains gram negative and is composed of peptidoglycan type A3ϒ enveloped by a layer of proteinaceous surface layer (S – layer) (5) | ||
==Ecology and Pathogenicity== | |||
Dehalobacter spp. are found in anaerobic environments like deep soils, groundwater aquifers, and river sediments. They compete with methanogens and iron reducers found in their habitat for hydrogen and other vital nutrients. All known Dehalobacter strains are non pathogenic. | |||
==Growth, Isolation and Cultivation== | |||
Dehalobacter restrictus is obligate anaerobic chemolithoheterotrophs that use chlorinated aliphatic and aromatic compounds as electron acceptors via reductive dehalogenation and hydrogen as the sole electron donor. Acetate serves as a carbon source. It was first isolated from an anaerobic fluidized bed reactor dechlorinating PCE (5). | |||
In a lab setting, Dehalobacter restrictus is either grown in anaerobic batch microcosms or a chemostat using a highly reduced medium. Generally, isolates are fed with pure dihydrogen as electron donor and a chlorinated compound of interest (8). (e.g. Tetrachloroethylene (PCE), 1,1,2 Trichloroethylene (TCE), Chloroform (CF), 1,1,1 Trichloroethane (1,1,1 TCA), DCM). Because of their restricive nutrional needs, isolates are hard to maintain and grow. As a result, Dehalobacter restrictus is often grown as mixed cultures consisting of acidogens and methanogens in order to supplement nutritional requirements (8). Another advantage of maintaining Dehalobacter in such a consortium is that electron donors other than hydrogen (e.g. Ethanol, lactate) can be fermented to yeild hydrogen as well as acetate which is then used up by Dehalobacter restrictus (8). Nevertheless, the choice of electron donors and accpetors are based on specific strains. Growth is sensitive to temperature and pH. The optimum ranges are 25.0 - 35.0 ºC and 6.8 to 7.6). pH is regulated using 0.1 M KOH or NaOH (8). | |||
==Metabolism== | |||
Dehalobacter restrictus has unique metabolic pathways that make them vital for bioremediation of several key recalcitrant environmental contaminants (4). | |||
===Reductive dehalogenation=== | |||
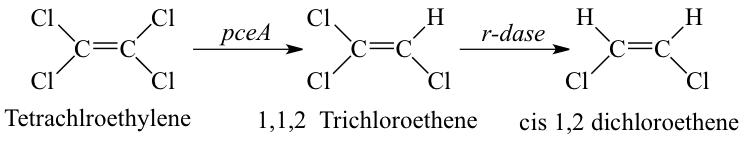
[[Image:Reductive_dehalogenation.png|frame|left|Metabolic pathway associated with organohalide respiration of Tetrachloroethylene mediated by Dehalobacter spp. resulting in incomplete reductive dechlorination to cis-1,2 dichlorethene (an environmentally unacceptable endpoint). From [Source, Hollinger et al., Arch Microbiol, 1998.]]] Reductive dehalogenation is the reduction of halogenated aliphatic and aromatic compounds using hydrogen as an electron donor. Microbially mediated reductive dehalogenation linked to growth is referred to as organohalide respiration. In addition to Dehalobacter restrictus, numerous microbial genera mediate organohalide respiration (e.g. Dehalococcoides, Dehalogimonas, etc.). When a chlorinated molecule is used as electron acceptor for organohalide respiration, and it is thus termed as microbially mediated reductive dechlorination. In this process, the chlorine atom on a chlorinated molecule is replaced by a hydrogen atom using a reductive dehalogenase enzyme (rdase) like pceA. | |||
Dehalobacter restrictus accepts chlorinated solvents, PCE and TCE, as electron acceptors. However, reductive dechlorination is incomplete and stops at cis-1,2 dichlorethene (cDCE). | |||
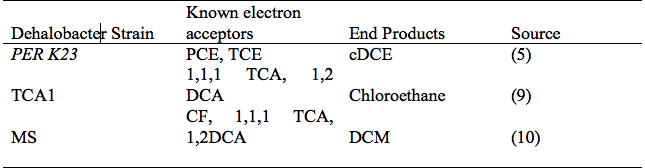
[[Image:Table_of_dehalobacters.png|frame|below|Key Dehalobacter spp. strains, known electron acceptors and end products of reductive dechlorination.]] | |||
Mixed populations of Dehalobacter spp. have also been shown to mediate reductive dehalogenation of 1,2 dichlorobenzene, monochlorobenzene, polychlorinated biphenyl and other recalcitrant chlorinated compounds. | |||
===Fermentation of Dichloromethane=== | |||
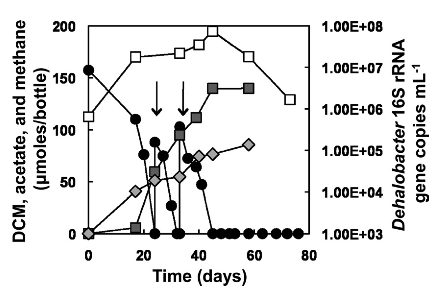
[[Image:Fermentation_of_dichloromethane.png|frame|below|Fermentative dehalogenation of DCM and a consequent increase in 16S rRNA gene copies of Dehalobacter gene copies. From [Justicia-Leon S.D. et al., 2012, AEM]]] Several different strains of Dehalobacter spp. have been known to degrade CF, and 1,1,1 TCA resulting in DCM or chloroethane, these end products often persisted and did not undergo further reductive dechlorination. However, a novel enrichment culture derived from pristine river sediment revealed another key dehalogenation pathway associated with Dehalobacter spp. This enrichment culture degraded DCM using fermentative dehalogenation. The resulting end products were acetate, methane, and chloride, which are environmentally benign. | |||
As | ==Significance== | ||
PCE and TCE are the predominant contaminants at hazardous waste sites in the United States (12). Although less prevalent, dichloromethane (DCM) is also found at a number of sites. Occasionally, the two types of contaminants are found comingled. PCE, TCE and DCM are excellent organic solvents and were used widely in dry cleaning, paint stripping and degreasing after 1960 (13). As of January 2000, the Environmental Protection Agency (EPA) classified PCE and DCM as likely to be human carcinogens by all routes of exposure while TCE is classified as a carcinogenic to humans by all routes of exposure (13). Studies indicate that chronic exposure to DCM can increase the probability of getting liver and lung cancer and benign growths in humans. The maximum contaminant level goal (MCLG) and the maximum contaminant level (MCL) for all three contaminants have been set to 0 and 0.005 mg/L, respectively (14). | |||
Bioremediation has emerged as a vital cost effective tool in the clean up of such hazardous waste sites. Monitored Natural Attenuation (MNA) resulting from bacterial dechlorinating consortia in the groundwater community is often used as a strategy for cleaning up chlorinated solvent plumes in ground water aquifer. On the other hand, mixed microbial containing microbes like Dehalobacter spp. are added to the groundwater aquifers along with an electron donor in order to induce reductive dechlorination in a process called as bioaugmentation (14). Because of their unique metabolic properties Dehalobacter spp. are uniquely suited for bioaugmentation at sites where PCE, TCE and DCM comingle. As a result several commercial vendors have developed trademarked reductive dechlorinating cultures that contain several different strains of Dehalobacter and Dehalococcoides spp. (4, 8). | |||
== | ==Author== | ||
This page was majorly revised and reorganized by Yogendra Kanitkar and Zhiguo Liu | |||
==References== | |||
== | 1. [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC1694251/ A 1,1,1-trichloroethane-degrading anaerobic mixed microbial culture enhances biotransformation of mixtures of chlorinated ethenes and ethanes. Appl. Environ. Microbiol. 72:7849-7856.] | ||
2. [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC1352275/ Growth of Dehalobacter and Dehalococcoides spp. during degradation of chlorinated ethanes. Appl. Environ. Microbiol. 72:428-436.] | |||
3. [http://aem.asm.org/content/78/4/1288.long Dichloromethane Fermentation by a Dehalobacter sp in an Enrichment Culture Derived from Pristine River Sediment. Applied and Environmental Microbiology 78:1288-1291.] | |||
4. [http://pubs.acs.org/doi/abs/10.1021/es403582f Bioaugmentation with Distinct Dehalobacter Strains Achieves Chloroform Detoxification in Microcosms. Environ Sci Technol 48:1851-1858.] | |||
5. [http://link.springer.com/article/10.1007/s002030050577 Dehalobacter restrictus gen. nov. and sp. nov., a strictly anaerobic bacterium that reductively dechlorinates tetra- and trichloroethene in an anaerobic respiration. Archives of Microbiology 169:313-321.] | |||
6. [http://download.springer.com/static/pdf/883/art%253A10.1007%252FBF00115297.pdf?auth66=1398023712_4144abe6a09060f6e5faf6e94e2379c0&ext=.pdf Isolation of an anaerobic bacterium which reductively dechlorinates tetrachloroethene and trichloroethene. Biodegradation 7:507-511.] | |||
7. [http://aem.asm.org/content/75/8/2400 A Novel Dehalobacter Species Is Involved in Extensive 4,5,6,7-Tetrachlorophthalide Dechlorination. Applied and Environmental Microbiology 75:2400-2405.] | |||
8. [http://www.ncbi.nlm.nih.gov/pubmed/19521729 Large-scale production of bacterial consortia for remediation of chlorinated solvent-contaminated groundwater. J Ind Microbiol Biot 36:1189-1197.] | |||
[http:// | 9. [http://www.sciencemag.org/content/298/5595/1023.abstract Microbial dehalorespiration with 1,1,1-trichloroethane. Science 298:1023-1025.] | ||
[http:// | 10. [http://onlinelibrary.wiley.com/doi/10.1111/j.1462-2920.2009.02150.x/abstract Chloroform respiration to dichloromethane by a Dehalobacter population. Environmental Microbiology 12:1053-1060.] | ||
[http://www.ncbi.nlm.nih.gov/ | 11. [http://www.ncbi.nlm.nih.gov/pubmed/24501624 Complete genome sequence of Dehalobacter restrictus PER-K23(T). Stand Genomic Sci 8:375-388.] | ||
[http:// | 12. [http://pubs.acs.org/doi/pdf/10.1021/es00080a600 Can contaminated aquifers at Superfund sites be remediated. Environmental Science and Technology 24:1464-1466.] | ||
[http:// | 13. [http://www.atsdr.cdc.gov/spl/ CERCLA priority list of hazardous substances. Agency for Toxic Substances and Disease Registry. ] | ||
[http://www. | 14. [http://www.epa.gov/osw/hazard/correctiveaction/resources/guidance/rem_eval/protocol.pdf Technical protocol for evaluating natural attenuation of chlorinated solvents in ground water EPA/600/R-98/128. U.S. Environmental Protection Agency, Office of Research and Development, Washington, DC.] | ||
Latest revision as of 20:03, 18 April 2014
A Microbial Biorealm page on the genus Dehalobacter
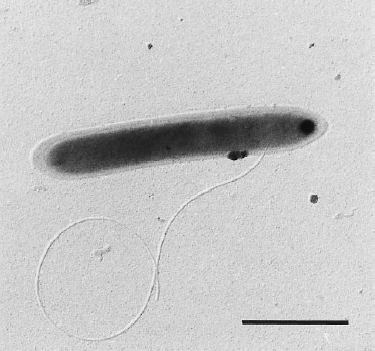
Classification
Higher order taxa:
Bacteria; Firmicutes; Clostridia; Clostridiales; Peptococcaceae; Dehalobacter
Species:
Dehalobacter restrictus PER K23, Dehalobacter sp. E1, Dehalobacter sp. TCA1, Dehalobacter sp. MS Dehalobacter sp. WL
|
NCBI: Taxonomy Genome |
Description and Significance
Dehalobacter restrictus is a rod shaped gram-negative obligate anaerobe found in deep soil sediments and groundwater aquifers and is phylogenetically related to several halorespiring genera like Desulfitobacterium and Desulfosporlosinus. It is a typical representative of Dehalobacter spp., which are capable of reductive dechlorination of chlorinated ethenes and chlorinated ethanes (1, 2) and fermentative dehalogenation of Dichloromethane (DCM) (3). These metabolic pathways make them relevant to bioremediation of ground water aquifers contaminated with chlorinated solvents (4). To date, several strains of Dehalobacter have been isolated (1, 3, 5-7)."
Genome Structure
The whole genome of Dehalobacter restrictus PER- K23 was recently sequenced revealing critical insights into phylogeny and metabolic capabilities of the genus. The 2,943 kbp long genome contains 2,826 protein coding and 82 RNA genes, including 5 16S rRNA genes. Interestingly, the genome contains 25 predicted rdase genes, the majority of which appear to be full length. The rdase genes are mainly located in two clusters, suggesting a much larger potential for organohalide respiration than previously anticipated (11).
Cell Structure
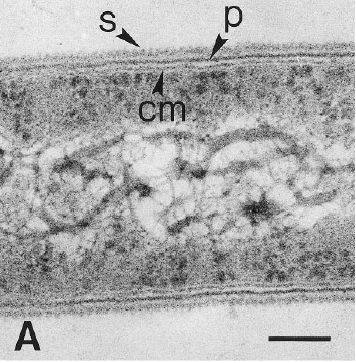
Dehalobacter restrictus cells are rod shaped, 2-3 µm in length and 0.3-0.5 µm in diameter with tapering ends. There is a single lateral flagellum at one end indicating that the cells are motile. The cells do not form spores under substrate and temperature induced stress. Cell wall stains gram negative and is composed of peptidoglycan type A3ϒ enveloped by a layer of proteinaceous surface layer (S – layer) (5)
Ecology and Pathogenicity
Dehalobacter spp. are found in anaerobic environments like deep soils, groundwater aquifers, and river sediments. They compete with methanogens and iron reducers found in their habitat for hydrogen and other vital nutrients. All known Dehalobacter strains are non pathogenic.
Growth, Isolation and Cultivation
Dehalobacter restrictus is obligate anaerobic chemolithoheterotrophs that use chlorinated aliphatic and aromatic compounds as electron acceptors via reductive dehalogenation and hydrogen as the sole electron donor. Acetate serves as a carbon source. It was first isolated from an anaerobic fluidized bed reactor dechlorinating PCE (5). In a lab setting, Dehalobacter restrictus is either grown in anaerobic batch microcosms or a chemostat using a highly reduced medium. Generally, isolates are fed with pure dihydrogen as electron donor and a chlorinated compound of interest (8). (e.g. Tetrachloroethylene (PCE), 1,1,2 Trichloroethylene (TCE), Chloroform (CF), 1,1,1 Trichloroethane (1,1,1 TCA), DCM). Because of their restricive nutrional needs, isolates are hard to maintain and grow. As a result, Dehalobacter restrictus is often grown as mixed cultures consisting of acidogens and methanogens in order to supplement nutritional requirements (8). Another advantage of maintaining Dehalobacter in such a consortium is that electron donors other than hydrogen (e.g. Ethanol, lactate) can be fermented to yeild hydrogen as well as acetate which is then used up by Dehalobacter restrictus (8). Nevertheless, the choice of electron donors and accpetors are based on specific strains. Growth is sensitive to temperature and pH. The optimum ranges are 25.0 - 35.0 ºC and 6.8 to 7.6). pH is regulated using 0.1 M KOH or NaOH (8).
Metabolism
Dehalobacter restrictus has unique metabolic pathways that make them vital for bioremediation of several key recalcitrant environmental contaminants (4).
Reductive dehalogenation
Reductive dehalogenation is the reduction of halogenated aliphatic and aromatic compounds using hydrogen as an electron donor. Microbially mediated reductive dehalogenation linked to growth is referred to as organohalide respiration. In addition to Dehalobacter restrictus, numerous microbial genera mediate organohalide respiration (e.g. Dehalococcoides, Dehalogimonas, etc.). When a chlorinated molecule is used as electron acceptor for organohalide respiration, and it is thus termed as microbially mediated reductive dechlorination. In this process, the chlorine atom on a chlorinated molecule is replaced by a hydrogen atom using a reductive dehalogenase enzyme (rdase) like pceA.
Dehalobacter restrictus accepts chlorinated solvents, PCE and TCE, as electron acceptors. However, reductive dechlorination is incomplete and stops at cis-1,2 dichlorethene (cDCE).
Mixed populations of Dehalobacter spp. have also been shown to mediate reductive dehalogenation of 1,2 dichlorobenzene, monochlorobenzene, polychlorinated biphenyl and other recalcitrant chlorinated compounds.
Fermentation of Dichloromethane
Several different strains of Dehalobacter spp. have been known to degrade CF, and 1,1,1 TCA resulting in DCM or chloroethane, these end products often persisted and did not undergo further reductive dechlorination. However, a novel enrichment culture derived from pristine river sediment revealed another key dehalogenation pathway associated with Dehalobacter spp. This enrichment culture degraded DCM using fermentative dehalogenation. The resulting end products were acetate, methane, and chloride, which are environmentally benign.
Significance
PCE and TCE are the predominant contaminants at hazardous waste sites in the United States (12). Although less prevalent, dichloromethane (DCM) is also found at a number of sites. Occasionally, the two types of contaminants are found comingled. PCE, TCE and DCM are excellent organic solvents and were used widely in dry cleaning, paint stripping and degreasing after 1960 (13). As of January 2000, the Environmental Protection Agency (EPA) classified PCE and DCM as likely to be human carcinogens by all routes of exposure while TCE is classified as a carcinogenic to humans by all routes of exposure (13). Studies indicate that chronic exposure to DCM can increase the probability of getting liver and lung cancer and benign growths in humans. The maximum contaminant level goal (MCLG) and the maximum contaminant level (MCL) for all three contaminants have been set to 0 and 0.005 mg/L, respectively (14). Bioremediation has emerged as a vital cost effective tool in the clean up of such hazardous waste sites. Monitored Natural Attenuation (MNA) resulting from bacterial dechlorinating consortia in the groundwater community is often used as a strategy for cleaning up chlorinated solvent plumes in ground water aquifer. On the other hand, mixed microbial containing microbes like Dehalobacter spp. are added to the groundwater aquifers along with an electron donor in order to induce reductive dechlorination in a process called as bioaugmentation (14). Because of their unique metabolic properties Dehalobacter spp. are uniquely suited for bioaugmentation at sites where PCE, TCE and DCM comingle. As a result several commercial vendors have developed trademarked reductive dechlorinating cultures that contain several different strains of Dehalobacter and Dehalococcoides spp. (4, 8).
Author
This page was majorly revised and reorganized by Yogendra Kanitkar and Zhiguo Liu
References
9. Microbial dehalorespiration with 1,1,1-trichloroethane. Science 298:1023-1025.
11. Complete genome sequence of Dehalobacter restrictus PER-K23(T). Stand Genomic Sci 8:375-388.
13. CERCLA priority list of hazardous substances. Agency for Toxic Substances and Disease Registry.