Bacteroide composition in the gut: Difference between revisions
| Line 10: | Line 10: | ||
Recently, genomic and proteomic analyses have provided insight into the mechanisms in which microbiota adapt and thrive within varying microenvironments. Sequencing B. thetaiotaomicron and B. fragilis show notable genomic commonalities. These Bacteroides, like eukaryotes, show low gene content for the size of their genome (Xu and Gordon, 2003). A large portion of the their genetic make-up consists of proteins containing over 1,000 amino acids, which, it seems, allow for a vast selection of relevant proteins for gene expression. Putative relevant proteins have been determined by homology to other known proteins. Of the 4779 proteins identified in the B. thetaiotaomicron proteome, 58% were ascribed a supposed function based on other known proteins, 18% were homologous to proteins without a known function, and 24% were not homologous to any protein identified in the public domain (Xu et al., 2003). The variety of functions is presumed to aid Bacteroides species in multiple ways: the proteome induces the mechanism by which some Bacteroides acquire and hydrolyze otherwise indigestible dietary polysaccharides, and includes an environment-sensing apparatus with extracytoplasmic function sigma factors and one- and two-component signal transduction systems (Xu et. al., 2003). | Recently, genomic and proteomic analyses have provided insight into the mechanisms in which microbiota adapt and thrive within varying microenvironments. Sequencing B. thetaiotaomicron and B. fragilis show notable genomic commonalities. These Bacteroides, like eukaryotes, show low gene content for the size of their genome (Xu and Gordon, 2003). A large portion of the their genetic make-up consists of proteins containing over 1,000 amino acids, which, it seems, allow for a vast selection of relevant proteins for gene expression. Putative relevant proteins have been determined by homology to other known proteins. Of the 4779 proteins identified in the B. thetaiotaomicron proteome, 58% were ascribed a supposed function based on other known proteins, 18% were homologous to proteins without a known function, and 24% were not homologous to any protein identified in the public domain (Xu et al., 2003). The variety of functions is presumed to aid Bacteroides species in multiple ways: the proteome induces the mechanism by which some Bacteroides acquire and hydrolyze otherwise indigestible dietary polysaccharides, and includes an environment-sensing apparatus with extracytoplasmic function sigma factors and one- and two-component signal transduction systems (Xu et. al., 2003). | ||
<i>B. thetaiotaomicron</i> contains the capacity to utilize glycans derived from the colon. The majority (61%) of the necessary enzymes (glycohyrolases) potentially exist in the periplasm, and are necessary to the polysaccharide fermentation process that increases energy availability to the host. Other extracellular proteins have been shown to facilitate the binding of starches to the cell surface to be digested by other extracellular enzymes (Xu et al., 2003). | |||
Additionally, B. thetaiotaomicron was shown to contain a significant population of ECF-sigma factors, which according to other identified homologous genes, are cotranscribed with regulatory proteins that respond to external environmental stimulus and act on gene expression accordingly. | Additionally, B. thetaiotaomicron was shown to contain a significant population of ECF-sigma factors, which according to other identified homologous genes, are cotranscribed with regulatory proteins that respond to external environmental stimulus and act on gene expression accordingly. | ||
Revision as of 22:14, 21 March 2015
The Bacterial kingdom has played a significant role in Homo sapien evolution. Only ten percent of the cells existing in the body are human. Genetically, we are about 1% human, and 99% bacterial (Xu & Gordon, 2003).
The human colon contains the majority of microorganisms in the body, and 25% of these are species of Bacteroides. Bacteroides species are anaerobic, non-spore forming, gram-negative rods that have adapted to, and now thrive in, the human gut(Wexler, 2007). The relationship between Bacteroides and the host has recently been considered mutual, given that the relationship increases the fitness of both species. Given the long history of coevolution between microbiota and the intestine, it has now been shown that bacteroides function as a multifunctional organ that provides metabolic components we do not contain in our own genome (Xu, 2003). Some of these metabolic traits include carbohydrate fermentation, freeing simplified carbohydrates to be reabsorbed by the large intestine and to be used for energy by the host. Through various metabolic mechanisms, Bacteroides provide simplified amino acids and vitamins, while simultaneously utilizing a wide range of dietary polysaccharides for growth (Hooper, Midtvedt, and Gordon. 2002).
Due to the metabolic role of Bacteroides in carbohydrate fermentation, there has been some evidence showing a correlation between diet, gut flora, and obesity. Given the significant role Bacteroides play in the gut, the effect of diet on their mechanisms of action and composition overall is a relevant topic of analysis (Turnbaugh, 2009). How body flora composition might affect adiposity is particularly crucial in light of the drastic shift in food source in the United States and its consequent growing obesity epidemic.
Genomic and Proteomic Advances
Recently, genomic and proteomic analyses have provided insight into the mechanisms in which microbiota adapt and thrive within varying microenvironments. Sequencing B. thetaiotaomicron and B. fragilis show notable genomic commonalities. These Bacteroides, like eukaryotes, show low gene content for the size of their genome (Xu and Gordon, 2003). A large portion of the their genetic make-up consists of proteins containing over 1,000 amino acids, which, it seems, allow for a vast selection of relevant proteins for gene expression. Putative relevant proteins have been determined by homology to other known proteins. Of the 4779 proteins identified in the B. thetaiotaomicron proteome, 58% were ascribed a supposed function based on other known proteins, 18% were homologous to proteins without a known function, and 24% were not homologous to any protein identified in the public domain (Xu et al., 2003). The variety of functions is presumed to aid Bacteroides species in multiple ways: the proteome induces the mechanism by which some Bacteroides acquire and hydrolyze otherwise indigestible dietary polysaccharides, and includes an environment-sensing apparatus with extracytoplasmic function sigma factors and one- and two-component signal transduction systems (Xu et. al., 2003).
B. thetaiotaomicron contains the capacity to utilize glycans derived from the colon. The majority (61%) of the necessary enzymes (glycohyrolases) potentially exist in the periplasm, and are necessary to the polysaccharide fermentation process that increases energy availability to the host. Other extracellular proteins have been shown to facilitate the binding of starches to the cell surface to be digested by other extracellular enzymes (Xu et al., 2003).
Additionally, B. thetaiotaomicron was shown to contain a significant population of ECF-sigma factors, which according to other identified homologous genes, are cotranscribed with regulatory proteins that respond to external environmental stimulus and act on gene expression accordingly.
The majority of one-component signal transduction systems were shown to be similar to other genes responsible for nutrient utilization, providing further evidence of Bacteroides’ genetically evolved fitness advantage in nutrient acquisition.
Through genomic and proteomic sequencing, some of the mechanism underlying the adaptive capabilities of the Bacteroide species in the human gut have been identified. The expansive array of proteins available to them mediates their ability to respond to their environment, thrive within the rapidly changing microenvironment of the colon, and acquire and ferment complex carbohydrates for their own energy source as well as for the host.
Double brackets: [[
Filename:1369260_bild1_b--ckhed.jpg
Thumbnail status: |thumb|
Pixel size: |300px|
Placement on page: |right|
Legend/credit: Bacteroides thetaiotaomicron in relation to food particles in the mouse intestine, provided by Larsson et al. (2012) in the study
Analysis of gut microbial regulation of host gene expression along the length of the gut and regulation of gut microbial ecology through MyD88.
Closed double brackets: ]]
Other examples:
Bold
Italic
Subscript: H2O
Superscript: Fe3+
Overall paper length should be 3,000 words, with at least 3 figures with data.
Enterotype and Diet
In 2011, Peer Bork and colleagues identified three clusters of microbiota species, termed enterotypes, and defined as “densely populated areas in a multi-dimensional space of community composition” (p. 177). Type 1 is characterized by high levels of Bacteroides, type two is dominated by Prevotella, and Ruminococcus is prevalent in type 3. In accordance with other genomic and proteomic analysis, the Bacteroides and Parabacteroides found in the type 1 enterotype fermented carbohydrates and proteins by the use of galactosidases, hexosaminidases, proteases enzymes for degradation as well as through glycolysis and pentose phosphate pathways. It was shown that enterotype composition is species driven, however, identification of molecular functioning of each species is necessary to determine health differences between hosts with varying enterotypes. Age, body mass index, gender, or nationality showed no clear causation to enterotype composition.
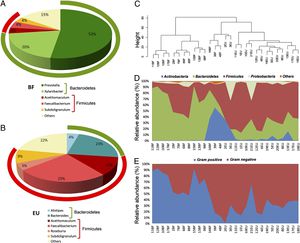
16S rRNA gene surveys reveal a clear separation of two children populations investigated. (A and B) Pie charts of median values of bacterial genera present in fecal samples of BF and EU children (>3%) found by RDP classifier v. 2.1. Rings represent corresponding phylum (Bacteroidetes in green and Firmicutes in red) for each of the most frequently represented genera. (C) Dendrogram obtained with complete linkage hierarchical clustering of the samples from BF and EU populations based on their genera. The subcluster located in the middle of the tree contains samples taken from the three youngest (1–2 y old) children of the BF group (16BF, 3BF, and 4BF) and two 1-y-old children of the EU group (2EU and 3EU). (D) Relative abundances (percentage of sequences) of the four most abundant bacterial phyla in each individual among the BF and EU children. Blue area in middle shows abundance of Actinobacteria, mainly represented by Bifidobacterium genus, in the five youngest EU and BF children. (E) Relative abundance (percentage of sequences) of Gram-negative and Gram-positive bacteria in each individual. Different distributions of Gram-negative and Gram-positive in the BF and EU populations reflect differences in the two most represented phyla, Bacteroidetes and Firmicutes. Legend taken directly from De Fillipe et al., 2010.
However, long-term dietary changes have been shown to alter the microbiota of the gut. A study by Wu and colleagues (2011) analyzed fecal samples, using diet inventories and 16S rDNA sequencing, from 98 individuals in order to determine the effect of diet on enterotype identity and microbiome composition. The authors concluded that while microbiome composition changed within 24 hours, enterotype identity shifts are associated with long-term dietary changes. Composition of enterotypes, particularly protein and animal fat (Bacteroides) versus carbohydrates (Prevotella), were most affected in long-term analysis. Their findings provide evidence that diet affects the amount of bacteroides living in the gut, and that the change is time-dependent.
Another study also showed a possible correlation between long-term diet and enterotype composition. De Filippo et al., in 2010, used high-throughput 16S rDNA sequencing and biochemical analyses to assess microbiota differences between children eating a fiber-rich, high-carbohydrate diet in Burkina Faso and European children fed diets high in fat and protein. The European children showed high levels of Bacteroides, while the Burkina Faso children contained microbiomes enriched by the Prevotella enterotype, known to contain genes that allow for xylan and cellulose hydrolysis. This genetic advantage may be completely lacking in children fed western diets. While there are many distinguishing factors between the two populations that might account for enterotype differences, diet is an obvious possible causative factor given what is known about the metabolic properties of bacterial species in the gut.
The authors of the Africa/Europe comparison hypothesize that diseases associated with enterotype composition may be treated by long-term dietary changes. It is also suggested that preservation of human communities in isolated areas is important, in that they may contain contain fitness advantages due to ancient coevolutionary symbioses between themselves and their microbiomes.
Section 3
Include some current research in each topic, with at least one figure showing data.
Further Reading
[Sample link] Ebola Hemorrhagic Fever—Centers for Disease Control and Prevention, Special Pathogens Branch
References
Edited by Maggie Schein, a student of Suzanne Kern in BIOL168L S2 (Microbiology) in The Keck Science Department of the Claremont Colleges Spring 2014.