Introduction to Organisms: Difference between revisions
No edit summary |
|||
| Line 274: | Line 274: | ||
==References== | ==References== | ||
1. Sylvia, D. M., Fuhrmann, J.F., Hartel, P.G., and D.A Zuberer (2005). Principles and Applications of Soil Microbiology. New Jersey: Pearson Education Inc. | |||
2. Ingham, Elaine R. Ingham R. "Chapter 4: SOIL FUNGI." Soil Biology. Natural Resources Conservation Service Illinois, n.d. Web. 9 Feb. 2016 | |||
3. Reid, Greg, and Percy Wong. "SOIL BACTERIA AND FUNGI - NEW SOUTH WALES." Soil Bacteria and Fungi. New South Wales Department of Primary Industries, 2005. Web. 11 Feb. 2016. | |||
4. "Fungi." Microbiology Online. Microbiology Society, n.d. Web. 11 Feb. 2016. | |||
5. Tortora, G.J., Funke, B.R., Case, C.L. (2012). “Microbiology an Introduction.” 11th ed. Pearson Benjamin Cummings, San Francisco. | |||
6. Rizzo, D. “Intro to Fungi.” Hutchinson Hall UC Davis, Davis. 28 Sept. 2015. Lecture. | |||
7. Rizzo, D. “Ascomycota Part I.” Hutchinson Hall UC Davis, Davis. 28 Sept. 2015. Lecture. | |||
8. Rizzo, D. “Basidiomycota Part I.” Hutchinson Hall UC Davis, Davis. 28 Sept. 2015. Lecture. | |||
9. Pegg G.F., Brady B.L. “Verticillium Wilts.” New York, CABI Publishing 2002. Google Books. Web. 11 Feb. 2016. http://books.google.com | |||
10. Hebert, Paul. "What Is a Protist / What Are Protozoa? - Encyclopedia of Life." Encyclopedia of Life. N.p., n.d. Web. 11 Feb. 2016.http://eol.org/info/456 | |||
11. "MicrobeWorld." Protista. N.p., n.d. Web. 11 Feb. 2016. http://www.microbeworld.org/types-of-microbes/protista | |||
12. Madigan, M.T., Martinko, J.M. (2006). Brock Biology of Microorganisms. New Jersey: Pearson Prentice Hall. | |||
13. Roesch, L.F., Fulthorpe, R. R., Riva, A., Casella, G., Hadwin, A. K. M., Kent, A. D., Daroub, S. H.; Camargo, F. A. O.; Farmerie, W. G.; Triplett, E. W.(2007). Pyrosequencing enumerates and contrasts soil microbial diversity. ISME Journal, 1(4).283-290. | |||
14. Amann, R.I, Ludwig, W., Schleifer, K.H.(1995).Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiology and Molecular Biology Reviews. 59(1). 143-169. | |||
15. Geers, D., Cohan, F.M., Lawrence J.G., Spratt, B.G., Coenye, T. Feil, E.J., Stackebrandt, E., Van de Peer, Y., Vandamme, P., Thompson, F.L., Swings, J. (2005). Opinion: Re-evaluating prokaryotic species. Nature Reviews Microbiology. 3(9). 733-739. | |||
16. Patzelt, D.J., Hodac, L., Friedl, T., Pietrasiak, N., Johansen, J.R. (2014). Biodiversity of soil cyanobacteria in the hyper-arid Atacama Desert, Chile. Journal of phycology. 50(4). 698-710. | |||
17. Makhalanyane, T. P., Valverde, A., Velázquez, D., Gunnigle, E., Van Goethem, M. W., Quesada, A., & Cowan, D. A. (2015). Ecology and biogeochemistry of cyanobacteria in soils, permafrost, aquatic and cryptic polar habitats. Biodiversity and Conservation, 24(4), 819-840. | |||
18. Corliss, John. "Protist | Biology." Encyclopedia Britannica Online. Encyclopedia Britannica, n.d. Web. 23 Feb. 2016. <http://www.britannica.com/science/protist>. | |||
19. Ekelund, Flemming; Saj, Stephane; Vestergård, Mette; Bertaux, Joanne; Mikola, Juha. (2009). “The “soil microbial loop” is not always needed to explain protozoan stimulation of plants”. Soil Biology & Biochemistry. Vol. 41 Issue 11, p2336-2342, 7p. | |||
20. Bonkowski, Michael. "Protozoa and Plant Growth: The Microbial Loop in Soil Revisited." New Phytologist New Phytol 162.3 (2004): 617-31. Web. | |||
21. Hoorman, James. "The Role of Soil Protozoa and Nematodes." Fact Sheet (n.d.): n. pag. Ohio State University, 2011. Web. | |||
22. Corliss, John. "Protist | Biology." Encyclopedia Britannica Online. Encyclopedia Britannica, n.d. Web. 23 Feb. 2016. <http://www.britannica.com/science/protist | |||
23. Lynn, Denis H(Apr 2012) Ciliophora. In: eLS. John Wiley & Sons Ltd, Chichester. http://www.els.net [doi: 10.1002/9780470015902.a0001966.pub3] | |||
24. "flagellate". Encyclopædia Britannica. Encyclopædia Britannica Online. Encyclopædia Britannica Inc., 2016. Web. 23 Feb. 2016 <http://www.britannica.com/science/flagellate>. | |||
25. "Mastigophora." World of Microbiology and Immunology. 2003. Encyclopedia.com. 23 Feb. 2016<http://www.encyclopedia.com>. | |||
26. “Plant Life." : Euglenoids. N.p., n.d. Web. 23 Feb. 2016. <http://lifeofplant.blogspot.com/2011/04/euglenoids.html> | |||
27. Parales, R.E. and Martin, M. (2015) Microbial Diversity Laboratory Manual, MIC 105L, Winter 2015. Department of Microbiology and Molecular Genetics, College of Biological Sciences, The University of California, Davis. | |||
28. Arnold, Paul. "What Does an Amoeba Eat? Plus Other Ameoba Info and Questions." Bright Hub. N.p., n.d. Web. 04 Mar. 2016. | |||
29. "MicrobeWorld." Slime Molds. N.p., n.d. Web. 24 Feb. 2016. <http://www.microbeworld.org/types-of-microbes/protista/slime-molds> | |||
30. Leeuwenhoek., A. V. (2009, November 18). One gram of soil: A microbial biochemical gene library. Retrieved March 13, 2016, from http://www.ncbi.nlm.nih.gov/pubmed/19921459 | |||
31. Coleman, D. C., Crossley, D. A. & Hendrix, P. F. "Soil food webs: Detritivory and microbiology in soils," in Fundamentals of Soil Ecology, 2nd ed. (Elsevier, 2004) 236-241. | |||
32. Welcome to Agriinfo.in. (n.d.). Retrieved February 21, 2016, from http://agriinfo.in/?page=topic&superid=5&topicid=172 | |||
33. Gouthro, T., & Rice, D. (1987, December 15). Interaction Terminology. Retrieved February 21, 2016, from http://www.rpi.edu/dept/chem-eng/Biotech-Environ/MixCul/ecodefn.htm | |||
34. AMENSALISM. (n.d.). Retrieved February 21, 2016, from https://biostuds.wikispaces.com/AMENSALISM | |||
35. Natural Resources Conservation Service. (n.d.). Retrieved March 10, 2016, from | |||
http://www.nrcs.usda.gov/wps/portal/nrcs/detailfull/soils/health/biology/?cid=nrcs142p2_053865 | |||
36. Ecological Linkages Between Aboveground and Belowground Biota. (n.d.). Retrieved March 10, 2016, from http://science.sciencemag.org/content/304/5677/1629 | |||
37. Gottesman, S., Harwood, C. S., Schneewind, O., Offre, P., Spang, A., & Schleper, C. (n.d.). Annual review of microbiology. Retrieved March 9, 2016, from http://eebweb.arizona.edu/faculty/saleska/SWES.410.510/Readings/Offre_Spang_Schleper_2013_AnnRevMicro_Archaea_Biogeochemistry.pdf | |||
38. Kingdom Archaea. (n.d.). Retrieved March 10, 2016, from http://plantphys.info/organismal/lechtml/archaea.shtml | |||
39. What are Archaea? - Encyclopedia of Life. (n.d.). Retrieved March 10, 2016, from http://eol.org/info/457 | |||
40. Flagellum. (n.d.). Retrieved March 10, 2016, from http://www.newworldencyclopedia.org/entry/Flagellum#Archaeal_flagellum | |||
41. Archaea: Morphology. (n.d.). Retrieved March 10, 2016, from http://www.ucmp.berkeley.edu/archaea/archaeamm.html | |||
42. Gottesman, S., Harwood, C. S., Schneewind, O., Offre, P., Spang, A., & Schleper, C. (n.d.). Annual review of microbiology. Retrieved March 9, 2016, from http://eebweb.arizona.edu/faculty/saleska/SWES.410.510/Readings/Offre_Spang_Schleper_2013_AnnRevMicro_Archaea_Biogeochemistry.pdf | |||
43. Archaea. (n.d.). Retrieved March 10, 2016, from http://www.eoearth.org/view/article/150172/ | |||
44. Binary Fission: Cell Division & Reproduction of Prokaryotes. (n.d.). Retrieved March 10, 2016, from http://www.scienceprofonline.com/microbi | |||
45. Bates, S. T., Berg-Lyons, D., Caporaso, J. G., Walters, W. A., Knight, R., & Fierer, N. (2010, November 18). Examining the global distribution of dominant archaeal populations in soil. Retrieved February 21, 2016, from http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3105767/ | |||
46. Scow, K. M. "Lecture 1 Part 1." Lecture, Davis. | |||
47. Rizzo, D. "Lecture 1." Lecture, Davis. | |||
48. Rizzo, D. "Lecture 3." Lecture, Davis. | |||
49. C.J. Alexopoulos & C. W. Mims, Introductory Mycology 3rd edition. 1979. Wiley NY. Web accessed Mar 8, 2016 from http://www.ucmp.berkeley.edu/fungi/fungimm.html | |||
50. Rizzo, D. "Lecture 2." Lecture, Davis. | |||
51. Ingham, E. “Chapter 4: Soil Fungi” (2016) Oregon State University. Retrieved Feb. 20, 2016, from http://extension.illinois.edu/soil/SoilBiology/fungi.htm | |||
52. Keller, N., Turner, G., Bennett, J. “Fungal Secondary Metabolism- from Biochemistry to Genomics.” 2005 Nature Reviews Microbiology p/ 937 to 947. | |||
Revision as of 02:08, 15 March 2016
Introduction
The soil ecosystem in an important environment that allows both plants and animals to survive, grow and reproduce. The significance of the soil as an ecosystem is at times overlooked despite its major contributions to the environment. The soil environment is a complex and varied microbial habitat.[1] The plants and organisms inhabiting the soil contribute to its thriving diversity. Soil life is diverse in morphology, metabolism, size, and many other characteristics. It is believed that “there is sufficient [microbial] DNA in 1 g of soil to extend 1,598km.” [30] Soil is a heterogeneous environment with various zones of rhizosphere, aggregates, organic matter and animal residues.[31] Given each zone is a small component of the larger soil ecosystem, rhizosphere accounts for only 5 to 7 percent of the soil environments but contains over “70 percent of the bacterial- and fungal-feeding nematodes.”[31]
Since, all three domains of life: archaea, bacteria and eukarya, inhabit the soil environment it is difficult to observe each microbe individually within the soil ecosystem.Only a few representative groups of each domain will be discussed to provide examples of the roles these organisms play in soil. This page attempts to outline the major microbial groups within the diverse soil environment.The interactions between soil microbes amongst the soil microbial diversity. The concept of survival of the fittest is seen in the soil food web, which allows only a selective few with the correct nutrients to thrive in a soil environment. The interactions of soil microbes is not only a part of nature but it also only selects those organisms that are fit for survival, growth and reproduction. The soil food web and symbiotic relationships are used to explain the dynamic interactions between soil organisms.
Soil Food Web and Microbial Interactions
To survive in a soil environment, an organism must be able to adapt to changing environments and organism interactions. The interactions seen between organisms allows only the fittest to survive, thrive, grow and reproduce in the soil. The only type of interaction shown in (Figure 1.1) is predation. Other common interactions in the soil environment are:
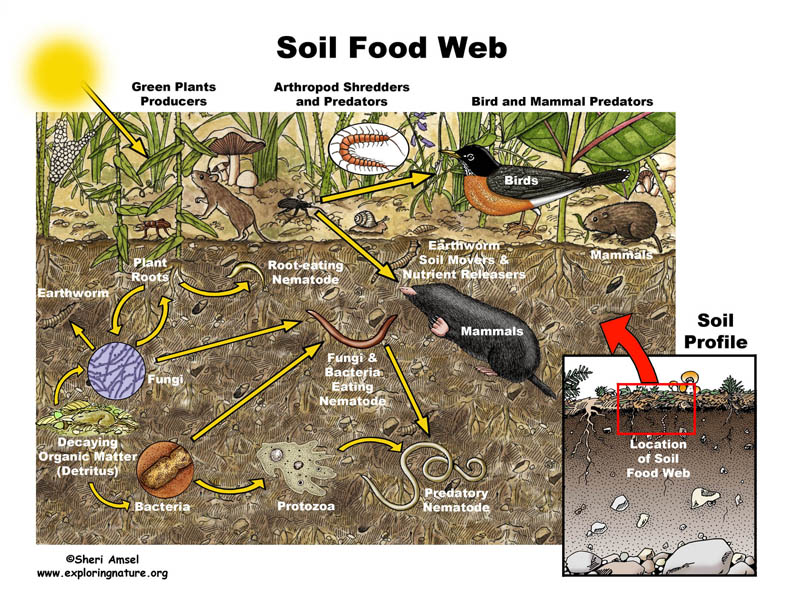 Figure 1.1 Soil Food Web. From[1]
Figure 1.1 Soil Food Web. From[1]
I. Commensalism: Organism (A) benefits from the presence of Organism (B) while (B) is unaffected by the presence of (A).[32]
II. Mutualism: Organism (A) benefits from Organism (B) and Organism (B) benefits from Organism (A) and becomes a necessary association for each other’s survival. [32] [33]
III. Synergism: Organism (A) benefits from Organism (B) and Organism (B) benefits from Organism (A) but the relationship is not necessary for survival.[33]
IV. Competition: Depending on the organisms this interaction benefits organisms who can readily adapt to the environment and overcome any obstacles needed to survive, grow and reproduce.[33]
V. Amensalism: An interaction in which Organisms (A) and (B) develop a partnership where (B) is unaffected by (A) but organism (A) is negatively affected by (B).[34]
VI. Parasitism: Organism (A) benefits from Organism (B) but Organism (B) does not benefit positively from Organism (A).
VII. Predation: “An interaction between organisms in which one benefits and one is harmed based on the ingestion of the smaller sized organism, the prey, by the larger organism, the predator.”[33]
The soil food web consists of microbes which play an important role in maintaining the complexity of the soil environment. The interactions observed in the food web are not only based on predation, which directly affects the nutrient cycling and degradation of pollutants, but also the formation of aggregates.[35] The production of specific nutrients, such as ammonium created by microbial metabolic functions, helps to enrich the the soil for plant use.[35] Soil organism activity, along with the presence of soil organic matter, create stabilizing aggregates.[35] The hyphae within the soil environment and the microbes help form large, stable aggregates in the soil habitat.[35] The biodiversity in the soil plays a pivotal role in degrading pollutants, and also prevents pathogens from inhabiting the soil environment through competition.[35] The food web in the soil environment is necessary to maintain the soil nutrients, biological activity, and detoxifying pollutants. Thus, it directly maintains cultivation, productiveness and prosperity of the soil.
In (Figure 1.1), all three domains, Archaea, Bacteria, and Eukarya, are represented in the soil food web. If one takes a closer look at the figure, the soil food web is not only dependent on small and large microbes but also dead organic matter and plant roots. Dead and living matter in the soil environment contribute to maintaining the food web. Plant residues also are apart of the soil food web. They contribute to the soil’s carbon source.[35] The plant residues either, release carbon dioxide to the environment, or contribute to the soil organic matter. Soil organic matter contributes to soil’s richness and helps to form aggregates.[35] The main focus in the image above is the diversity of organisms, both in domains and in size. The larger organisms are usually the predators. The smaller organisms eventually fall prey, due to their limited mobility, respiration, and size. The eukarya and small bacteria are apparent figures seen in the figure above, but archaea also contribute to the food cycle as they break down ammonia for plants to use. [35]
The notable interactions of life that take place in the soil environment is evident both below-ground level, as seen in the figure 1.1, but also in the above-ground level.[36] Worms also impact the soil food web, acting as both drivers of the food web both belowground and aboveground.[36]
3 Domains
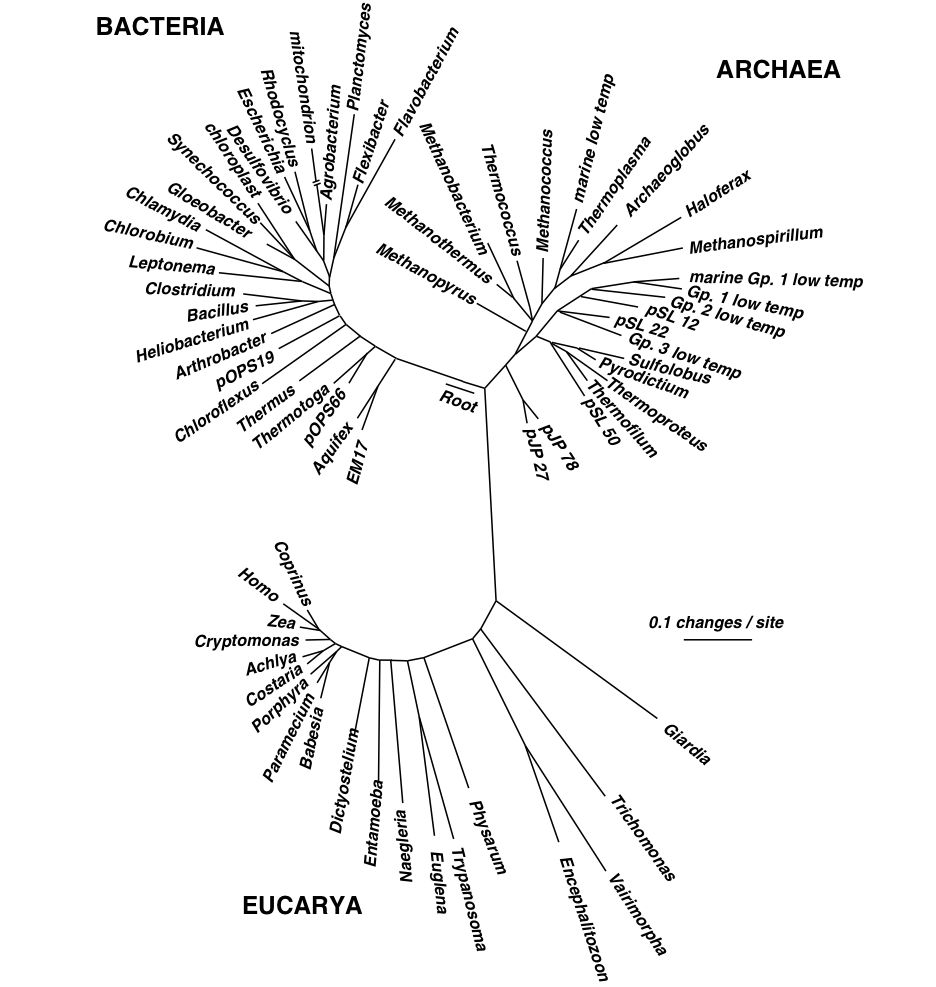
Carl Woese and the ssRNA

Carl Woese (Figure 1.3) was an American microbiologist famous for pioneering the technique of phylogenetic taxonomy through use of a 16S (for prokaryotic) and 18S (for eukaryotic) ribosomal RNA, also known as small subunit ribosomal RNA [46]. This refined Linnaean classification, taking out the guesswork classification by using a gene that was present in every living organism- the small subunit ribosomal RNA. This method of classification is comprehensive because all organisms possess this form of genetic material. Ssu rRNA is sequenced because the genes are highly conserved. Due to their functional constancy, some have variable regions to differentiate organisms, and are easy to sequence. This technique also elucidated the tree of life, which shows the three domains of microbial lineage diversity: Bacteria, Archaea, and Eukarya. DNA can be compared using a sample database that allows identification of unknown organisms to a species level.
Domain Bacteria
Introduction to Bacteria
Bacteria are single celled prokaryotic organisms that lack membrane bound organelles. They are involved in many important processes within the soil such as decomposition, bioremediation, symbiosis and biological transformation of nutrients. They are present in all terrestrial soil environments.[1] Soil is a heterogenous environment that varies in moisture content, temperature, oxygen requirements, carbon source and many other bacterial growth conditions. Those wide ranging growth factors account for the enormous diversity in soil bacteria populations. One gram of soil can contain between 26,000 to 52,000 different bacterial species.[13] In the entire microbial world, only about 1 percent of bacterial species can be cultured under laboratory conditions which leaves an immense number of bacterial species yet to be described. [14]
Classification based on phenotypic characteristics, such as cellular morphology, metabolism, gram staining, oxygen requirements, carbon source and other traits, was used more readily before modern DNA sequencing. But, phenotypic classification is still used and Bergey’s Manual of Systematic Bacteriology is the most widely used source for determining species based on characteristic species traits.
Traditional species definitions are not as useful in bacteria. OTUs, or operational taxonomic units, based on DNA sequencing similarity, are more applicable to bacteria. Typically, those bacteria with 97 percent similar DNA sequence are classified within the same OTU.[15] The 16S small subunit ribosomal RNA is used to substantiate phylogenetic relationships between different bacterial groups.
Morphology
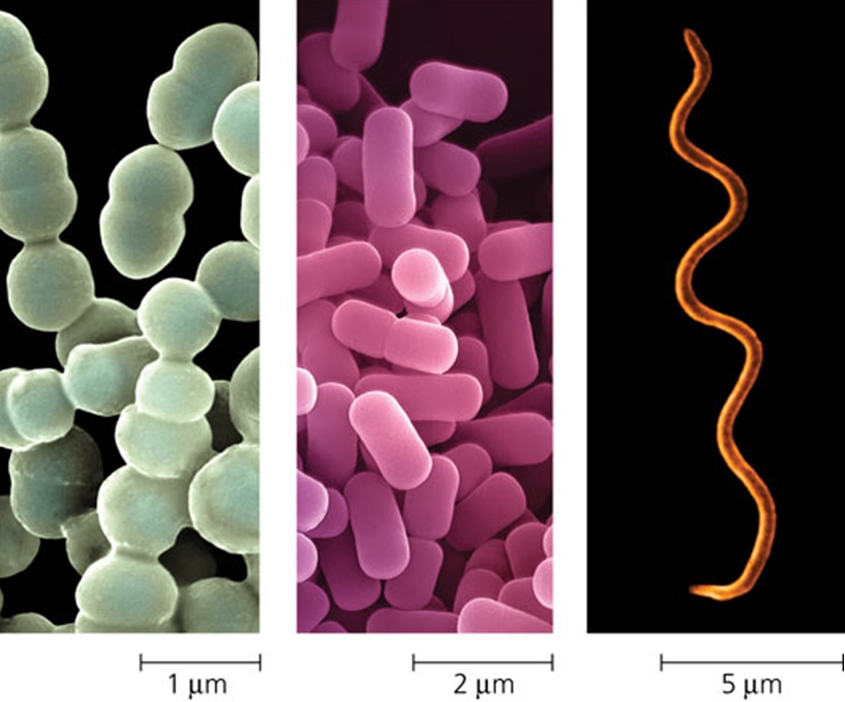
Bacterial cells measure approximately 1μm wide and 1-5μm long.[12] They are smaller than eukaryotic microbes.Due to their small size, bacteria can more easily adapt to changing soil environments than larger eukaryotes.[1] Most bacteria are singled celled, but actinomycetes grow in branching filaments. There are many different bacterial cell shapes. The most common are bacilli (rods), cocci (spheres), spirilla (spirals) and actinomycete (filaments).
Flagella - There are many different types and different arrangement of flagella. Movement in soil environments is dependent on soil moisture content. Though movement in soil is limited, flagellar function is important to allow bacteria to find more favorable microsites.[1]
Cell membrane - The cell membrane encloses the cytoplasm. It is semi-permeable phospholipid bilayer that regulates transport of nutrients into the cell and waste products out of the cell.
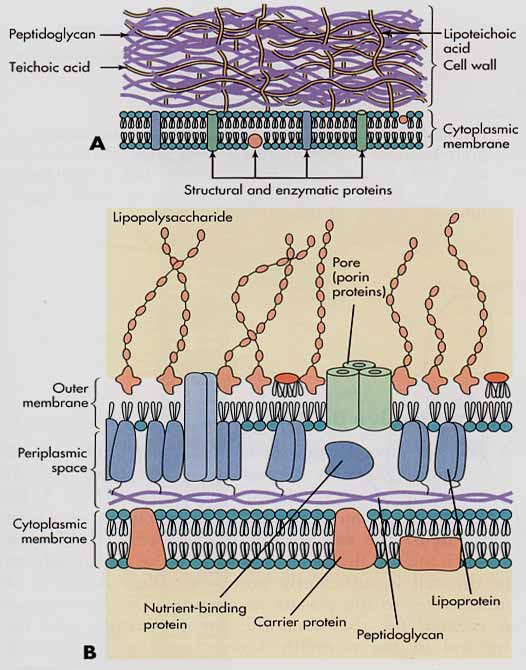
Cell wall - Most bacteria have a rigid cell wall surrounding the cell membrane which protects the cell from osmotic lysis. The cell wall is made of a crosslinked polysaccharide, called peptidoglycan, that is found only in bacteria.[1] The cell wall gives the cell shape and structure, but it is porous enough to permit small water soluble molecules to flow through to the cell membrane surface.[1]
- Gram positive bacteria have a thick cell wall made of several layers of peptidoglycan. The cell wall is the outermost structure, as they do not have an outer membrane. The thick cell wall structure make gram positive bacteria more resilient to osmotic shock. Gram positive bacteria can tolerate 5 to 10 times more turgor pressure than gram Gram negative bacteria.[1] Teichoic acids in the cell wall may also help to bind to cationic nutrients in soil surroundings.[1]
- Gram negative bacteria have a thin cell wall surrounded by an outer membrane made of phospholipids and lipopolysaccharides. Polysaccharides of the outer membrane interact with surrounding cations and are involved in attachment of bacteria to soil particles. [1] Periplasm is a gel like substance that occupies the space between the outer membrane and the cell wall of gram negative bacteria.[1] The outer membrane and periplasmic space is thought to provide protection from environmental toxins.[1] Proteins within the periplasm are thought to begin the process of catalyzing hydrolysis of organic and toxic substances.[1]
Endospores are made by Bacillus and Clostridium in response to environmental stress.[1] Endospores are extremely resistant structures that can withstand extreme temperatures up to 150℃.[12] They can also withstand extreme pressure, desiccation, radiation, pH and chemicals.[12] The endospore protects the bacterial DNA until environmental conditions are more favorable and germination can proceed. Soil is the most common habitat for endospore forming bacteria.[12]
Extracellular polymers - Many bacteria secrete a slime layer or capsule made of polysaccharides or glycoproteins.[1] The outer polysaccharide coating of bacteria helps in attachment to surfaces and the formation of biofilms.[1] Bacterial capsule secretions play a big role in soil aggregation and soil structure.[1] When the carbon to nitrogen ratio within soils is high, microbes secrete the excess carbon as extracellular polysaccharides.[1]
Reproduction
Most bacteria reproduce by binary fission.[12] The bacterial cell expands to twice the original cell size, duplicates the bacterial DNA and pinches off, producing two daughter cells. Each resultant daughter cell is a clone of the parent cell. Prokaryotic Archaea also reproduce by binary fission.
- Spore formation: Actinomycetes are a large group of filamentous bacteria, primarily found in soil.[1] Actinomycetes are morphologically similar to fungi. Actinomycetes form long filaments called hyphae. Unlike most bacteria, the majority of actinomycetes reproduce using asexual spores called conidia.[1] Though they share many characteristic with fungi, actinomycetes are still prokaryotes. They are smaller than fungi and the cell wall contains peptidoglycan.[1] Actinomycetes break down a wide variety of organic compounds and play an important role in organic matter decomposition. [1] Streptomyces, a subgroup within actinomycetes, produce geosmin, which gives soil the characteristic earthy smell.[12]
Metabolism
Along with archaea, bacteria are the most metabolically diverse and employ many different nutritional strategies.[1]
- Heterotrophs - Most bacteria are heterotrophic, obtaining carbon from preformed organic substances. [1]
- Autotrophs - Carbon dioxide serves as the main source of carbon. Some cyanobacteria, nitrifying and sulfur-oxidizing bacteria are autotrophs.[1]
- Organotrophs - Most bacteria obtain electrons from organic substances.[1]
- Lithotrophs - Some cyanobacteria, nitrifying bacteria and sulfur-oxidizing bacteria obtain electrons from inorganic substances.[1]
- Phototrophs - Cyanobacteria use light as the main source of metabolic energy.[1]
- Chemotrophs - All other nonphotosynthetic bacteria do not rely on light for metabolic energy.[1]
Contributions to the Cycle
Bacteria play a significant role in the cycling of a wide range of elements.
- Carbon Assimilation is the process of fixing carbon dioxide into organic forms. Carbon fixation is performed by autotrophic bacteria, such as cyanobacteria.[1] Chemoautotrophic nitrifying bacteria, such as Nitrobacter, Nitrosospira and Nitrosomonas, also fix carbon.[1] Sulfur oxidizing bacteria, such as Thiobacillus, fix carbon as well.
- Carbon Mineralization results from microbial decomposition of organic carbon compounds into inorganic forms. As the organic carbon substances, like plant matter, are decomposed, inorganic carbon dioxide is produced. Most bacteria are heterotrophs that perform carbon mineralization. When microorganisms die, they are decomposed by successive microbial generations. Peptidoglycan, the main component of bacterial cell walls, is utilized as a polysaccharide substrate by other heterotrophic soil microbes.[1]
- Methanogenesis - Methanogenic bacteria produce methane from the reduction of carbon dioxide. They are all strict anaerobes that use carbon dioxide for the carbon source.[1] They can all use hydrogen as a source of energy and electron donor. [1]
- Nitrogen fixation: Azotobacter is a free living nitrogen fixing bacteria.[1] Rhizobium lives in a mutualistic relationship with plants by fixing nitrogen within root the nodules of legumes.[1] Anabaena and Nostoc have specialized heterocyte structures where nitrogen fixation occurs.[1]
- Nitrification: Nitrifying bacteria, such as Nitrobacter, Nitrosospira, and Nitrosomonas, oxidize ammonium to nitrate.[1]
- Denitrification: Alcaligenes, Flavobacterium, and Pseudomonas are common soil denitrifying bacteria.[1]
Domain Archaea
Introduction to Archaea
Vladimir Vernadsky decided, in 1926, that only two domains of life existed on Earth, Bacteria and Eukarya.[37] About 50 years later in 1977, Carl Woese and George Fox theorized that part of the bacterial domain possed multiple dissimilarities from bacteria and recognized it to be another domain of life, now called Archaea.[38] Woese and Fox’s theory regarding the differences of archaea were confirmed in 1990’s through the use of 16S rRNA and 18S rRNA.[38]
Despite the strongly held belief, in the 1990s, that archaea only inhabited extreme environments, recent discoveries have suggested their presence in abundant numbers all throughout the Earth. It is suggested that Archaea play notable roles in maintaining nutrient cycles.[37] Various methods of research, such as cultivation, culture-independent techniques and isotope-based methods, have facilitated the study of domain Archaea in carbon and nitrogen cycling. [37]
The use of genomic and biochemical data has permitted the study of archaeal structure.[37] Use of these research methods has shown that domain archaea and domain bacteria differ in cell wall characteristics yet, share various ways to transfer genetic information by replication and transcription from their common ancestor, Eukarya.[37] As of 2012, 116 genera of Archaea have been observed which consists of 450 species. Most of the archaeal species in these genera remains uncultivated, known only through its gene sequences and molecular surveys.[37]
Morphology
Archaea are characterized by various cell sizes. They are most commonly found to be small organisms ranging from “0.1 to 15 μ diameter and up to 200 μ long.”[38] The shape of many archaea are similar to bacteria, including bacillus, cocci, spirilla and “plate-like forms.” [38] Their cell membranes have ether linked lipids with no periplasmic space, similar to gram-positive bacteria, and contain no intracellular organelles.[39]
Flagella: Most motile archaea inhabit a motor rotary system that allows it to move its flagella through the chemical gradient similar to bacteria.[39] Despite its similarity in function and source of energy required for motility, the flagella of both archaea and bacteria differ in structure.[39] Archaeal flagella gets the energy for its movement by ATP and is composed of dependent filaments that rotate singularly. [40] Archaeal flagella are known to be thinner than bacteria flagellin.[40] Since, the archaeal common ancestor is not bacteria but eukarya, convergent evolution is a plausible explanations for the differences between bacterial and archaeal flagellin. [40]
Membranes: Archaea have a cell membrane that protects the cytoplasm and the nucleic acids from the environment. The archaeal cell membrane has a lipid bilayer with glycol-ether lipids that are more stable than ester linkages and help some archaea that live in extreme environments to withstand such conditions.[39] Most archaeal species are surrounded by a cell wall which protects the cell membrane and maintains the archaeal structure.[41]
Reproduction
Archaeal organisms reproduce by binary fission since they do not have nucleases thus, the DNA chromosome in a Archaeal organism replicate, separate and create two daughter cells.[43] The process of binary fission can be seen in Figure (1.6) process begins the parent cell duplicating the DNA. The DNA material then goes to the sides of the cell where an invagination to the cell occurs allowing the cell to become two individual cells. Binary fission is a very fast reproduction process, that allows archaeal organisms to grow at an exponential rate.[44]

Metabolism
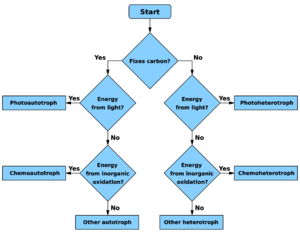
Archaea share similar metabolic characteristics to bacteria. The metabolic pathways taken by all archaea shown in the (Figure 1.7) demonstrates the great diversity and variation amongst archaea. [38] The kinds of archaea that are seen in the environment and the metabolic pathways taken by some can be summarized in the image.
- Photoautotrophs: use light as an energy source and carbon dioxide as the carbon source with the help of the Calvin Cycle.[38]
- Chemoautotrophs: use inorganic chemicals as their energy source and carbon dioxide as carbon source.
- Chemoheterotrophs: take organic chemicals as energy source and organic chemicals as carbon source. The pathways taken by chemoheterotrophs varies from respiration by the Krebs Cycle to fermentation. [38]
- Photoheterotrophs: use light as their energy source and organic chemicals as their carbon source.
Most archaea inhabiting in the soil environment are major contributors to the biogeochemical cycles.
Contributions to the Cycle
Archaea play a significant role in the nutrient cycles that take place in the ecosystem as they account for about 1-5% of all the prokaryotes present in the soil environment.[42]
Carbon Cycle: Archaea have an important role in the carbon cycle that takes place specifically in methane production, carbon mineralization and carbon assimilation all of all of which contribute to the soil’s carbon sink.[42] The carbon dense soil helps to make the soil organic matter rich and helps to form extracellular polymers which creates stable aggregates. For some microbes, an electron acceptor is need to carry out metabolic functions. [42] More info on Carbon Cycle.
- Carbon Assimilation: Involves the transformation of inorganic carbon into organic carbon compounds. This process is commonly done by autotrophic phyla Crenarchaeota, Thaumarchaeota, and Euryarchaeota that use carbon dioxide/ bicarbonate converting it to organic compounds.[42] Of the autotrophic lineages observed in carbon assimilation the Thaumarchaeota are strict autotrophs, the Euryarchaeota and Crenarchaeota are facultative autotrophs and the Halobacteriales and Thermococcales are obligate heterotrophs.[42]
- Carbon Mineralization: The process that converts organic carbon into the oxidized inorganic carbon, carbon dioxide is commonly done by both aerobes and anaerobes.[42] The heterotrophic microbes that use organic compounds as energy to oxidize the carbon source include, methanogens and methane-oxidizing archaea belonging to the phyla Euryarchaetota, and Crenarchaeota.[42]
- Methanogenesis: production of methane, a major greenhouse gas in the Earth’s atmosphere is commonly done by archaea upon reducing an oxidized compound.[42] The phyla Euryarchaeota is the only phyla that consists of archaea that produce methane all of which form five class: Methanopyri, Methanococci, Methanobacteria, Methanomicrobia and Thermoplasmata.[42] Methanogens are commonly found in anaerobic and aerobic conditions such as bodies of water, aerated soils and oxygenated waters.[42]
Nitrogen Cycle : Archaea play a pivotal role in the Nitrogen cycle and help create nitrogenous products for the plants and microbes. More info on Nitrogen cycle.
- Nitrogen Fixation: Fixing nitrogen from the environment and converting it to organic nitrogen in the soil or to inorganic nitrogenous form since both organic and inorganic products are in short supply in the biosphere and are fixed by many archaeal organisms.[42] N2 fixation by the environment is usually done by three major archaeal classes Methanobactera, Methanococci and Methanomicrobia.[42]
- Nitrification: Converting the ammonia (NH4+) in the soil to nitrite (NO2-) produces nitrate (NO3-) as an intermediate. [42] The transformation of ammonia to first nitrate then to nitrite is a two step process that is done by different microbes. [42] Most common organisms involved in nitrification include ammonia-oxidizing archaea which are lithotrophic organisms mostly, of the Thaumarchaeota phylum from the archaea domain.
- Denitrification : The conversion of atmospheric N2 using the electron acceptor NO3- or NO2- creating the formation of nitric oxide, nitrous oxide and/or N2.[42] The formation of the end product N2 may not always be produced but other intermediates NO and N2O, greenhouse gases may be produced.[42] Of the archaeal organisms that have been studied in denitrification process is done by Pyrobaculum aerophilum which accepts both NO3- and NO2- as an electron acceptor producing a variety of products NO2-, NO, N20 and N2.[42]
Domain Eukarya
Fungi
Introduction
Introduction
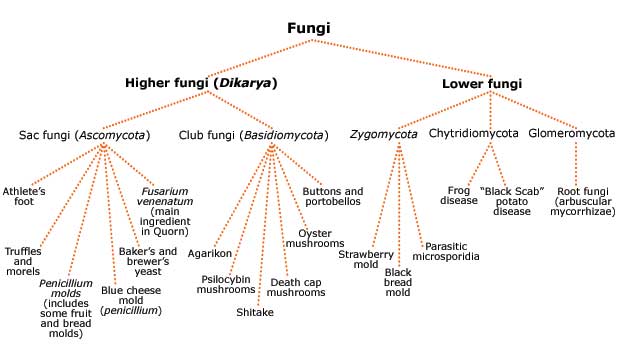
Fungi comes from the ancient Greek word “sphongos” meaning sponge in reference to the mushroom cap [47]. Fungi have the general characteristics of being eukaryotic, heterotrophic, external absorbers of nutrients, containing a hyphal thallus, reproducing via spore, and being pleomorphic (producing several forms). Fungi are generally organoheterotrophs that can survive in low pH and stay active at low moisture content[46]. Fungi reside in the domain Eukarya with an estimated species of 1.5 million, and less than five percent of them have ever been described. The general phyla included within Kingdom Fungi include Ascomycota, Basidiomycota, Glomeromycota, Zygomycota, Chytridiomycota, and Microsporidia [48].
Morphology
Morphology
Fungi can be either unicellular (yeasts) or multicellular. All multicellular fungi are made up of small, filamentous particles called hyphae. Some hyphae have cross walls called septa, while others do not- this is a large basis of fungal identification. Fungal cells generally have multiple nuclei. Fungal cell walls are generally composed of chitin, a long carbohydrate polymer that adds rigidity and structural support to fungi [49].
Reproduction
Reproduciton
Fungal reproduction can occur either sexually or asexually.
Sexual reproduction can occur by homothallic or heterothallic mycelia.
Asexual reproduction can occur by fragmentation, budding, or by producing spores (the most common method).
- Spores: spores come in many different forms. One of the most well known are conidiospores, which are unicellular or multicellular spores that get released from the tip of the hyphae.
Perfect fungi are those that can reproduce both sexually and asexually while imperfect fungi are those that can only reproduce asexually.
- Sexual reproduction always includes the stages plasmogamy (fusion of two haploid cells), karyogamy (creation of a diploid zygote), and meiosis (gametes are created).
Metabolism
Metabolism
Many different fungi display a myriad of metabolisms. There are primary metabolites that that help the fungi function properly, and secondary metabolites that are generally bioactive that are not the most pertinent to fungal growth (they can live without these). Secondary metabolites play a large role in chemistry today and can be manufactured into commonly used products such as new drugs. In general fungi are heterotrophic through release of extracellular enzymes into the neighboring environment to break complex molecules down into smaller ones. The materials are then absorbed and metabolized [50]. Fungi are sessile organisms that overcome their little to no movement through apical growth at the hyphal tip. Fungi are noted to grow almost everywhere on the Earth.
Soil Relevance
Soil Relevance
Fungi include some of the most abundant biomass of microorganisms in numerous soils. They play an important role as decomposers within the soil food web through conversion of difficult to digest organic material into forms many other organisms can use [51]. Their hyphae can bind soil particles together and create aggregates that increase soil water holding capacity. They perform important functions from water dynamics to nutrient cycling and disease suppression. Soil fungi can be categorized into three different groups based on their actions:
- decomposers: that convert dead organic material into fungal biomass
- mutualists: which colonize plant roots and help solubilize phosphorous and bring soil nutrients
- pathogens/ parasites: which help control diseases in soil via trapping organisms such as nematodes [51].
Fungi are seen less in agricultural soils due to tillage disrupting hyphae leading to less healthy soils. Fungi also play a very important role in the rhizosphere, the section of soil that is greatly influenced by the plant root system. This area of soil which is saturated with plant roots and fungal hyphae is known to support greater microbial activity.
Protista
Protists are mainly eukaryotic unicellular organisms. They used to belong to their own kingdom Protista, but are now known to be paraphyletic [18]. Protists are made up of protozoa, unicellular algae, and slime molds. The protozoa are divided into 4 major categories: the ciliates, the flagellates, the heliozoans, and the amoebas [10]. They are so small that they don’t need any specialized organelles in order to survive and can survive at low oxygen levels because of this. They are also known to use a contractile vacuole to remove excess water from their cells [10].
Protozoans
Protozoans
The microbial loop is a subsection of the microbial food web that accounts for the dissolved organic matter of systems. It’s suggested that Protozoa have an important role in the maintenance of soil nutrients, specifically nitrogen. A common known characteristic of protozoans is that they stimulate plant growth [19]. It’s hypothesized that they do this through the manipulation of nitrogen levels. In the presence of Protozoa, shoot biomass and shoot N levels are increased [20]. The protozoa is said to stimulate the mineralization of nitrogen via the microbial loop [20]. The indirect effects protozoans have on the nitrogen cycle are thought to be more important than the direct effects it has. This is because grazing stimulates microbial mineralization processes that in turn speeds up the nutrient cycles [20].
In general, Protozoans range from 5 to 500 µm in diameter, making them significantly larger than bacteria [21]. They are abundantly located near the surface of the soil (15cm deep) where roots are plentiful [21]. Most protozoans have two life cycles. One is their active form where they are motile and feed on prey. The other is the cyst form where they produce a thick coating to protect them from the environment and can go dormant for years.
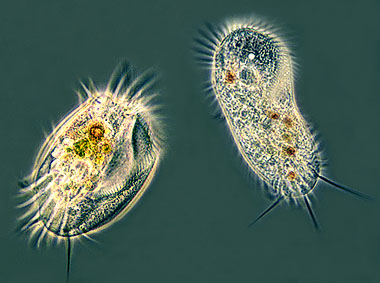
Ciliates
Cilliates: This group, within protozoans, is fairly diverse. A common feature shared amongst all of its members is the presence of at least one cilia at any stage of its development [10]. The cilia is used for movement and other things such as predation (tentacles). For movement, they use the massive amounts of tiny cilia to create wave like currents to propel the cell forwards. Most ciliates also have toxicysts which are used to capture prey [10]. Some ciliates even have a symbiotic relationship with algae living inside of them [10]. The algae use the CO2 produced by ciliates and turn it into O2 that can then be reused by the ciliates [10]. They are found in a wide array of habitats ranging from freshwater and soil to the rumen of Animals [22]. They are present in almost anywhere that contains water [23]. They are unicellular heterotrophs so they feed on bacteria and algae in their environment. Ciliates are one of the top predators in the microbial food web and are hypothesized to have been a major predatory group before animals evolved [23]. Parasitic ciliates exist that can cause the death of animals and are increasingly becoming dangerous in water [23].
Flagellates
Flagellates are one of the most commonly known protists. They are unicellular organisms that may be solitary, colonial, free living, or parasitic [24]. The hallmark feature of protists is that at some point in their life cycle, they possess a flagella. A flagellum is a long hair like structure that is used for locomotion, sensation, and sometimes predation [24]. The flagellum makes waves by whiplike lashing movements which propels the cell forwards. Flagellates also have a protective coating in the form of a thin pellicle to protect them from the environment [24].
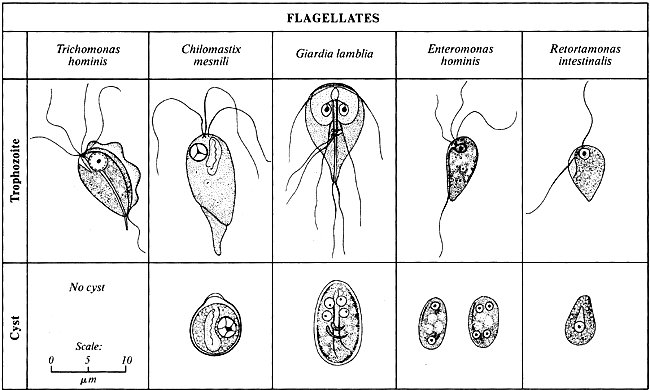
- Mastigophora:
Mastigophora is a subphylum of flagellates. Being a member of the flagellates, their key feature is the flagellum. Their habitats include freshwater, soil, and can act as animal parasites as well [25]. They use pseudopodia, a long outward protrusion, to trap and funnel food from the environment into their cell bodies [25]. The Mastigophora are mainly known for their parasitism, which can take place in both humans and animals [25]. Typanosomes are an example of Mastigophora which causes African sleeping sickness [25]. Giardia lamblia is another example that resides in the intestinal tract of warm-blooded animals and inflicts intestinal giardiases [25]. It is also a major contaminant of drinking water because it is resistant to chlorine, which is the most common water purifier [25]. Having a dormant cyst form also adds to its survival potential through filtration efforts [25].
- Euglenids:
Euglenids are one of the best known flagellates. They are found in freshwater and soil, as long as there are organic materials. They also participate in some symbiotic relationships with marine animals. Euglenid cells do not contain cell walls or other rigid membranes [26]. This gives the cell tremendous flexibility and allows it to change its shape to easily navigate through muddy terrain. Euglenids have two flagella, one which creates locomotion and movement for the cell [26]. The other flagellum does not protrude from the cell and is contained in the cell body [26].
Amoebas
Amoebas
Amoebae are single celled eukaryotes present in many different types of environments. They are scattered throughout a variety of eukaryotic lineages and therefore are not monophyletic [27]. Amoebae range in size from 5 micrometers to 3 mm and don’t have a defined shape, commonly recognized for being polymorphic. They are motile through the use of pseudopodia, which stretch and pull the cell along with it. The pseudopodia are also used for feeding. They are able to sense prey that can be engulfed and engulf them via phagocytosis. The amoeba’s main prey consists of bacteria, algae, and other protozoa [28]. The prey is moved around within the cell via food vacuoles, a feature of amoebae [28]. Another key characteristic of amoebae are the large contractile vacuoles inside the cell. The purpose of the contractile vacuole is to osmoregulate by pumping excess water out of the cell. In times of turmoil, amoebae have an encysted form that helps them get through the tough times. This form is useful when there is low nutrient availability to limit energy consumption. There are two main forms an amoeba can be characterized as. They can either be testate or nontestate. Testate amoeba produce shells, or “tests”, that help identify their species since each species has a unique shell.

Slime Molds
Slime molds are fungus like organisms that were at once classified as fungi, but have now been placed in the protozoan phylogeny. Slime molds are single cells that can live independently, but aggregate to form multicellular reproductive structures when necessary [29]. The fruiting body releases spores which germinate into new cells. Slime molds feed off of bacteria and are most commonly found in forests. They feed off of decaying vegetation as well so they can be seen on rotting logs and leaves.
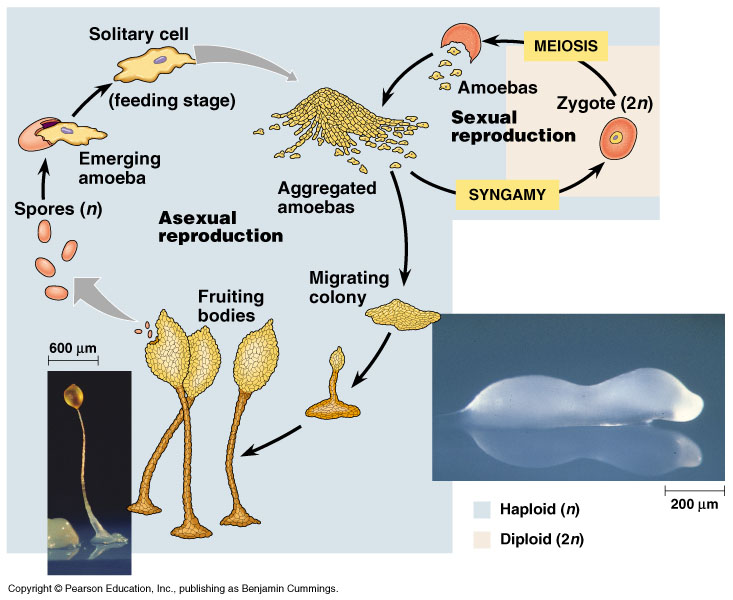
Current Research
Bacteria: To showcase the great diversity of soil bacterial environments, cyanobacteria have been studied in both desert and polar regions. Cyanobacteria can survive in extreme conditions likely because of their long evolutionary past.[16] Dr. Patzelt and his colleagues studied the diverse cyanobacteria residing in the Atacama desert, one of the driest parts of the world. The diverse taxa found in the Atacama desert can withstand prolonged periods of extreme desiccation.[16] The cyanobacteria dominate the soil crusts, affecting soil stability, soil structure and soil fertility.[16] Cyanobacteria inhabiting the Antarctic and Arctic regions, show similar adaptations to extreme environments as the cyanobacteria found in the desert. The polar cyanobacteria are particularly adapted to high altitudes.[17] They can withstand high ultraviolet radiation, desiccation, and repeated freeze-thaw cycles.[17] The autotrophic cyanobacteria supports heterotrophic microbes, providing an important ecosystem service in an environment with low microbial activity.[17]
Archaea: It is observed that in 150 soil samples acquired from North and South America, which inhabited only archaeal and bacterial species, about 150,000 species were observed.[45] Of the 150,000 species observed over 2,500 were identified as archaea based on their ssu rRNA.[45] In this study, the archaeal species were further analyzed to understand their role in the soil environment. Of the two common archaeal species, Crenarchaeota and Euryarchaeota were found in the soil samples.[45] Bates stated that, “Across the non-experimental soils examined the soil C:N ratio was the single best predictor of archaeal relative abundances” due to their influences in the nutrient cycles.[45]
Fungi: Fungi have been in the public eye for a while now due to the many different types of secondary metabolites they produce. Many of these secondary metabolites have bioactive compounds that can inhibit any kind of growth from protozoa, bacteria, parasites, insects, and even other fungi. A large area of intrigue is currently investigating the uses of these chemicals for human interests in use from medicine to plant and animal toxins [52].
Protozoa: It’s known that Protozoa stimulate plant growth in some way. It’s suspected that the Protozoa take part in the microbial loop which is responsible for the nutrient production for plants [19]. However, new developments have shown that the Protozoa themselves are responsible for the nitrogen mineralization and that other bacteria have no effect on the plant’s nitrogen sources [19]. This would mean that the stimulation of plant growth via nitrogen is solely due to the Protozoa and that the microbial loop is irrelevant [19].
References
1. Sylvia, D. M., Fuhrmann, J.F., Hartel, P.G., and D.A Zuberer (2005). Principles and Applications of Soil Microbiology. New Jersey: Pearson Education Inc.
2. Ingham, Elaine R. Ingham R. "Chapter 4: SOIL FUNGI." Soil Biology. Natural Resources Conservation Service Illinois, n.d. Web. 9 Feb. 2016
3. Reid, Greg, and Percy Wong. "SOIL BACTERIA AND FUNGI - NEW SOUTH WALES." Soil Bacteria and Fungi. New South Wales Department of Primary Industries, 2005. Web. 11 Feb. 2016.
4. "Fungi." Microbiology Online. Microbiology Society, n.d. Web. 11 Feb. 2016.
5. Tortora, G.J., Funke, B.R., Case, C.L. (2012). “Microbiology an Introduction.” 11th ed. Pearson Benjamin Cummings, San Francisco.
6. Rizzo, D. “Intro to Fungi.” Hutchinson Hall UC Davis, Davis. 28 Sept. 2015. Lecture.
7. Rizzo, D. “Ascomycota Part I.” Hutchinson Hall UC Davis, Davis. 28 Sept. 2015. Lecture.
8. Rizzo, D. “Basidiomycota Part I.” Hutchinson Hall UC Davis, Davis. 28 Sept. 2015. Lecture.
9. Pegg G.F., Brady B.L. “Verticillium Wilts.” New York, CABI Publishing 2002. Google Books. Web. 11 Feb. 2016. http://books.google.com
10. Hebert, Paul. "What Is a Protist / What Are Protozoa? - Encyclopedia of Life." Encyclopedia of Life. N.p., n.d. Web. 11 Feb. 2016.http://eol.org/info/456
11. "MicrobeWorld." Protista. N.p., n.d. Web. 11 Feb. 2016. http://www.microbeworld.org/types-of-microbes/protista
12. Madigan, M.T., Martinko, J.M. (2006). Brock Biology of Microorganisms. New Jersey: Pearson Prentice Hall.
13. Roesch, L.F., Fulthorpe, R. R., Riva, A., Casella, G., Hadwin, A. K. M., Kent, A. D., Daroub, S. H.; Camargo, F. A. O.; Farmerie, W. G.; Triplett, E. W.(2007). Pyrosequencing enumerates and contrasts soil microbial diversity. ISME Journal, 1(4).283-290.
14. Amann, R.I, Ludwig, W., Schleifer, K.H.(1995).Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiology and Molecular Biology Reviews. 59(1). 143-169.
15. Geers, D., Cohan, F.M., Lawrence J.G., Spratt, B.G., Coenye, T. Feil, E.J., Stackebrandt, E., Van de Peer, Y., Vandamme, P., Thompson, F.L., Swings, J. (2005). Opinion: Re-evaluating prokaryotic species. Nature Reviews Microbiology. 3(9). 733-739.
16. Patzelt, D.J., Hodac, L., Friedl, T., Pietrasiak, N., Johansen, J.R. (2014). Biodiversity of soil cyanobacteria in the hyper-arid Atacama Desert, Chile. Journal of phycology. 50(4). 698-710.
17. Makhalanyane, T. P., Valverde, A., Velázquez, D., Gunnigle, E., Van Goethem, M. W., Quesada, A., & Cowan, D. A. (2015). Ecology and biogeochemistry of cyanobacteria in soils, permafrost, aquatic and cryptic polar habitats. Biodiversity and Conservation, 24(4), 819-840.
18. Corliss, John. "Protist | Biology." Encyclopedia Britannica Online. Encyclopedia Britannica, n.d. Web. 23 Feb. 2016. <http://www.britannica.com/science/protist>.
19. Ekelund, Flemming; Saj, Stephane; Vestergård, Mette; Bertaux, Joanne; Mikola, Juha. (2009). “The “soil microbial loop” is not always needed to explain protozoan stimulation of plants”. Soil Biology & Biochemistry. Vol. 41 Issue 11, p2336-2342, 7p.
20. Bonkowski, Michael. "Protozoa and Plant Growth: The Microbial Loop in Soil Revisited." New Phytologist New Phytol 162.3 (2004): 617-31. Web.
21. Hoorman, James. "The Role of Soil Protozoa and Nematodes." Fact Sheet (n.d.): n. pag. Ohio State University, 2011. Web.
22. Corliss, John. "Protist | Biology." Encyclopedia Britannica Online. Encyclopedia Britannica, n.d. Web. 23 Feb. 2016. <http://www.britannica.com/science/protist
23. Lynn, Denis H(Apr 2012) Ciliophora. In: eLS. John Wiley & Sons Ltd, Chichester. http://www.els.net [doi: 10.1002/9780470015902.a0001966.pub3]
24. "flagellate". Encyclopædia Britannica. Encyclopædia Britannica Online. Encyclopædia Britannica Inc., 2016. Web. 23 Feb. 2016 <http://www.britannica.com/science/flagellate>.
25. "Mastigophora." World of Microbiology and Immunology. 2003. Encyclopedia.com. 23 Feb. 2016<http://www.encyclopedia.com>.
26. “Plant Life." : Euglenoids. N.p., n.d. Web. 23 Feb. 2016. <http://lifeofplant.blogspot.com/2011/04/euglenoids.html>
27. Parales, R.E. and Martin, M. (2015) Microbial Diversity Laboratory Manual, MIC 105L, Winter 2015. Department of Microbiology and Molecular Genetics, College of Biological Sciences, The University of California, Davis.
28. Arnold, Paul. "What Does an Amoeba Eat? Plus Other Ameoba Info and Questions." Bright Hub. N.p., n.d. Web. 04 Mar. 2016.
29. "MicrobeWorld." Slime Molds. N.p., n.d. Web. 24 Feb. 2016. <http://www.microbeworld.org/types-of-microbes/protista/slime-molds>
30. Leeuwenhoek., A. V. (2009, November 18). One gram of soil: A microbial biochemical gene library. Retrieved March 13, 2016, from http://www.ncbi.nlm.nih.gov/pubmed/19921459
31. Coleman, D. C., Crossley, D. A. & Hendrix, P. F. "Soil food webs: Detritivory and microbiology in soils," in Fundamentals of Soil Ecology, 2nd ed. (Elsevier, 2004) 236-241.
32. Welcome to Agriinfo.in. (n.d.). Retrieved February 21, 2016, from http://agriinfo.in/?page=topic&superid=5&topicid=172
33. Gouthro, T., & Rice, D. (1987, December 15). Interaction Terminology. Retrieved February 21, 2016, from http://www.rpi.edu/dept/chem-eng/Biotech-Environ/MixCul/ecodefn.htm
34. AMENSALISM. (n.d.). Retrieved February 21, 2016, from https://biostuds.wikispaces.com/AMENSALISM
35. Natural Resources Conservation Service. (n.d.). Retrieved March 10, 2016, from http://www.nrcs.usda.gov/wps/portal/nrcs/detailfull/soils/health/biology/?cid=nrcs142p2_053865
36. Ecological Linkages Between Aboveground and Belowground Biota. (n.d.). Retrieved March 10, 2016, from http://science.sciencemag.org/content/304/5677/1629
37. Gottesman, S., Harwood, C. S., Schneewind, O., Offre, P., Spang, A., & Schleper, C. (n.d.). Annual review of microbiology. Retrieved March 9, 2016, from http://eebweb.arizona.edu/faculty/saleska/SWES.410.510/Readings/Offre_Spang_Schleper_2013_AnnRevMicro_Archaea_Biogeochemistry.pdf
38. Kingdom Archaea. (n.d.). Retrieved March 10, 2016, from http://plantphys.info/organismal/lechtml/archaea.shtml
39. What are Archaea? - Encyclopedia of Life. (n.d.). Retrieved March 10, 2016, from http://eol.org/info/457
40. Flagellum. (n.d.). Retrieved March 10, 2016, from http://www.newworldencyclopedia.org/entry/Flagellum#Archaeal_flagellum
41. Archaea: Morphology. (n.d.). Retrieved March 10, 2016, from http://www.ucmp.berkeley.edu/archaea/archaeamm.html
42. Gottesman, S., Harwood, C. S., Schneewind, O., Offre, P., Spang, A., & Schleper, C. (n.d.). Annual review of microbiology. Retrieved March 9, 2016, from http://eebweb.arizona.edu/faculty/saleska/SWES.410.510/Readings/Offre_Spang_Schleper_2013_AnnRevMicro_Archaea_Biogeochemistry.pdf
43. Archaea. (n.d.). Retrieved March 10, 2016, from http://www.eoearth.org/view/article/150172/
44. Binary Fission: Cell Division & Reproduction of Prokaryotes. (n.d.). Retrieved March 10, 2016, from http://www.scienceprofonline.com/microbi
45. Bates, S. T., Berg-Lyons, D., Caporaso, J. G., Walters, W. A., Knight, R., & Fierer, N. (2010, November 18). Examining the global distribution of dominant archaeal populations in soil. Retrieved February 21, 2016, from http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3105767/
46. Scow, K. M. "Lecture 1 Part 1." Lecture, Davis.
47. Rizzo, D. "Lecture 1." Lecture, Davis.
48. Rizzo, D. "Lecture 3." Lecture, Davis.
49. C.J. Alexopoulos & C. W. Mims, Introductory Mycology 3rd edition. 1979. Wiley NY. Web accessed Mar 8, 2016 from http://www.ucmp.berkeley.edu/fungi/fungimm.html
50. Rizzo, D. "Lecture 2." Lecture, Davis.
51. Ingham, E. “Chapter 4: Soil Fungi” (2016) Oregon State University. Retrieved Feb. 20, 2016, from http://extension.illinois.edu/soil/SoilBiology/fungi.htm
52. Keller, N., Turner, G., Bennett, J. “Fungal Secondary Metabolism- from Biochemistry to Genomics.” 2005 Nature Reviews Microbiology p/ 937 to 947.
