Bdellovibrio exovorus: Difference between revisions
| Line 41: | Line 41: | ||
[[File:Bdellovibrio_exovorus_chromosome_map.png|300px|thumb|right|Chromosome map of <I>Bd. exovorus</I> strain JSS.]] | [[File:Bdellovibrio_exovorus_chromosome_map.png|300px|thumb|right|Chromosome map of <I>Bd. exovorus</I> strain JSS.]] | ||
<B>Closest Relatives:</B> | |||
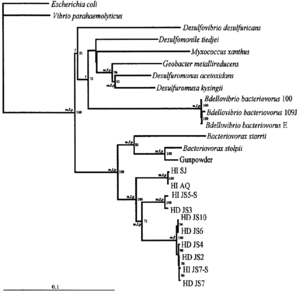
<I>Bdellovibrio exovorus</I> belongs to the major branch of prokaryotes of <I>deltaproteobacteria</I>. The most closely related species is <I>Bdellovibrio bacteriovorus</I>. Based on 16S rRNA sequences, the most closely related genera include <I>Desulfomonile, Desulfuromonas,</I> and more distantly, <I>Bacteriovorax.</I> | |||
<B>Extra-chromosomal Elements and Genetic Tools:</B> | |||
Both PCR and Southern blotting with a DIG-labelled probe at the <I>hit</I> locus (short for "host interaction"), a gene indicative of <I>bdellovibrio</I> species. | Both PCR and Southern blotting with a DIG-labelled probe at the <I>hit</I> locus (short for "host interaction"), a gene indicative of <I>bdellovibrio</I> species. | ||
| Line 50: | Line 52: | ||
Transposon mutagenesis has been used to identify genes used to attach to solid surfaces for predation. This includes the gene <I>bd3852</I>, which codes for the protein PilT2, an ATP-dependent motor that helps depolymerize the protein PilA, a component of type IV pili in host cells. | Transposon mutagenesis has been used to identify genes used to attach to solid surfaces for predation. This includes the gene <I>bd3852</I>, which codes for the protein PilT2, an ATP-dependent motor that helps depolymerize the protein PilA, a component of type IV pili in host cells. | ||
<B>Genome Sequencing:</B> | |||
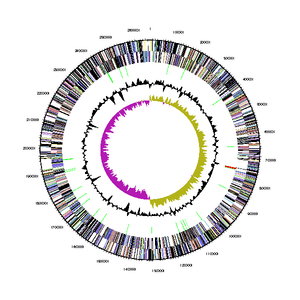
The genome sequence of <I>Bd. exovorus</I> was determined using whole genome sequencing (Weizmann Institute of Science, 2013), and can be found in its entirety on the [https://img.jgi.doe.gov/cgi-bin/m/main.cgi?section=TaxonDetail&page=taxonDetail&taxon_oid=2541047022 Joint Genome Institute] database. It has a total genome size of 2,657,893 bases, a %G+C content of 41.92%, and 2,656 genes. Of those genes, 2,619 (98.61%) are protein coding genes, 37 (1.39%) are RNA genes (of which 34 are for tRNA and 3 are for rRNA), and 1 (0.04%) is a pseudogene. Its 16S rRNA gene is 1502 bp long, and is only 90-93% similar to other strains of <I>Bd. bacteriovorus</I>. The bacterium produces a total of 2669 proteins, of which 130 are hydrolytic enzymes used to drain prey bacteria of their cytoplasmic contents. | |||
==Nutrition and metabolism== | ==Nutrition and metabolism== | ||
Revision as of 04:16, 3 April 2017
Classification
Higher order taxa
Domain: Bacteria
Phylum: Proteobacteria
Class: Deltaproteobacteria
Order: Bdellovibrionales
Family: Bdellovibrionaceae
Species
|
NCBI: Taxonomy Bdellovibrio exovorus |
Bdellovibrio exovorus
Description and significance
a. Bdellovibrio exovorus is a parasitic, predatory bacterium whose name means “Leech-like vibrating outside-devourer” (Gr. Bdella, leech, sucker; L. vibrio, vibrating; Gr. exo-, outside, L. -vorus, to eat.) (2). Bd. exovorus was first isolated from sewage in London, Ontario, Canada, and sequenced in 2013 by the Weizmann Institute of Science (3).
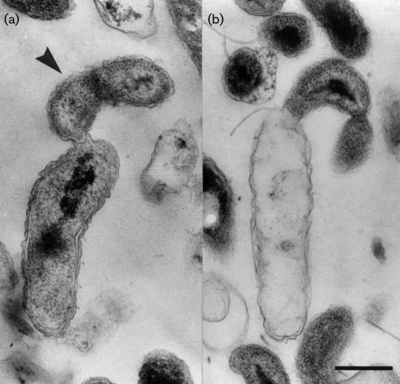
b. Bd. exovorus are gram-negative, comma-shaped rods, about 0.5 um wide and 0.5-1.4 um long. They have a single, polar, sheathed flagellum that is 29 nm wide. These obligate predators grow as small plaques on lawns of prey bacteria. Bd. exovorus' most common prey cell is Caulobacter crescentus, and actually will not prey upon Escherichia coli (2).
Existing genus MicrobeWiki Link: https://microbewiki.kenyon.edu/index.php/Bdellovibrio
Genome and genetics
Closest Relatives:
Bdellovibrio exovorus belongs to the major branch of prokaryotes of deltaproteobacteria. The most closely related species is Bdellovibrio bacteriovorus. Based on 16S rRNA sequences, the most closely related genera include Desulfomonile, Desulfuromonas, and more distantly, Bacteriovorax.
Extra-chromosomal Elements and Genetic Tools:
Both PCR and Southern blotting with a DIG-labelled probe at the hit locus (short for "host interaction"), a gene indicative of bdellovibrio species. The CRISPR system has been used to count DNA scaffolds in the genome.
Transposon mutagenesis has been used to identify genes used to attach to solid surfaces for predation. This includes the gene bd3852, which codes for the protein PilT2, an ATP-dependent motor that helps depolymerize the protein PilA, a component of type IV pili in host cells.
Genome Sequencing:
The genome sequence of Bd. exovorus was determined using whole genome sequencing (Weizmann Institute of Science, 2013), and can be found in its entirety on the Joint Genome Institute database. It has a total genome size of 2,657,893 bases, a %G+C content of 41.92%, and 2,656 genes. Of those genes, 2,619 (98.61%) are protein coding genes, 37 (1.39%) are RNA genes (of which 34 are for tRNA and 3 are for rRNA), and 1 (0.04%) is a pseudogene. Its 16S rRNA gene is 1502 bp long, and is only 90-93% similar to other strains of Bd. bacteriovorus. The bacterium produces a total of 2669 proteins, of which 130 are hydrolytic enzymes used to drain prey bacteria of their cytoplasmic contents.
Nutrition and metabolism
a.Describe the growth characteristics of your bacterial species; sources of C, N, electrons; respires/ferments, uses O2, etc.
Bdellovibrio exovorus is an obligate predator, gathering nutrients by feeding on other gram-negative bacterial species like Caulobacter crescentus and Stenotrophomonas maltophilia. However, it is somewhat selective in the organisms it preys upon, as it will not use species like Escherichia coli as a host cell, despite them also possessing gram-negative cell walls.
Life Cycle:
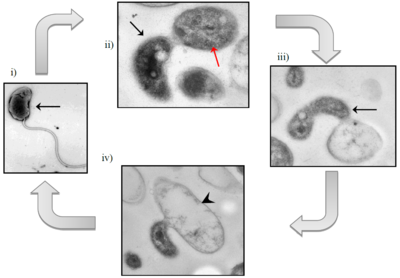
The life cycle of Bd. exovorus has two phases: a free-swimming attack phase and a stationary growth phase. In the first phase, <Bd. exovorus rapidly propels itself via its flagellum to randomly attack a prey cell, attaching to the exterior of its cell wall. In the second phase, its flagellum is lost and it becomes permanently attached to the host cell. Bd. exovorus releases hydrolytic enzymes into the host's cytoplasm, degrading its contents and transporting them back to the cell surface for Bd. exovorus to absorb. From here, Bd. exovorus can continue to grow and replicate, producing daughter cells while remaining attached to the cell. Bd. exovorus will continue elongation and binary fission until the host prey cell is completely emptied of its cytoplasmic structures and nutrients, making the prey into an empty "ghost cell."
All other known plaque-forming species within the family Bdellovibrionaceae are identified by their growth within the periplasmic space of their prey bacteria. However, Bd. exovorus has no periplasmic phase in its life cycle. Because Bd. exovorus does not inhabit the periplasmic space, it does not form the rounded bdelloplast out of the host cell that is common among members of the genus Bdellovibrio, or even related genera like Bacteriovorax, Bacteriolyticum, and Predibacter.
Replication in Bd. exovorus occurs via binary fission, producing two daughter cells that begin the attack phase.
Metabolism and Nutrients:
Bd. exovorus uses complete metabolic pathways like glycolysis and the TCA cycle, and can complete the pentose phosphate pathway non-oxidatively from D-ribose-5-phosphate. It undergoes complete amino acid biosynthesis for alanine, cysteine, aspartic acid, glutamic acid, glycine, glutamine, serine, threonine, tyrosine, and possibly lysine. It can undergo complete de novo nucleotide biosynthesis for dCTP and dTTP, and possibly for dATP and DGTP using inosine as a starting compound. Fatty acids can also be completely degraded and metabolized, presumably into acetyl-coA for use in the TCA cycle.
Bd. exovorus also contains cytochrome C within its electron transport chain, but it lacks catalase, superoxide dismutase, or peroxides needed to break down oxidative radicals.
b.What kinds of culture conditions (temp, pH, media) are needed for laboratory study?
Culture Conditions:
For laboratory study, cultures of Bd. exovorus are grown on minimal medium with the appropriate prey bacterium grown as a lawn. Bd. exovorus will grow as plaques on that bacterial lawn, leaving the empty husks of the prey cells behind.
One culturing method used by researchers involved growing Bd. exovorus on 1/10-strength yest extract-peptone medium supplemented with calcium, although dilute nutrient broth medium has also been used. HM buffer is often added to media, made of HEPES, CaCl2, and MgSO4. While optimal growth occurs around 30 °C, Bd. exovorus can be kept in a sterile tube at 4 °C for 1 month or stored long-term with 25% (w/v) glycerol at -80 °C. One research team calculated a 46.1 mol% G+C content in 1x SSC (Saline-Sodium Citrate buffer) using the equation [G+C=1.99(Tm-66.0)], so its genomic DNA must have undergone thermal denaturation at 89.2 °C.
c.What kinds of waste, by-products, volatile compounds are generated?
Waste and By-Products:
No waste, by-products, or volatile compounds appear to have been explicitly discussed in the literature regarding Bd. exovorus, although the ghost cells left behind after consuming the cytoplasmic contents of a host cell could be considered a waste product. Bd. exovorus also sheds its flagellum once entering the growth phase, so perhaps that could be considered a by-product as well.
Ecology / Pathology
Ecology: How is your microorganism important in the ecosystem where it is found? How does it impact other organisms in the environment (could be positive or negative impact)?
Ecology:
While bdellovibrios have been found ubiquitously in rivers, soil, compost, and wastewater,Bd. exovorus in particular was isolated from raw sewage, preying on other bacteria that flourish in that environment. Because it significantly reduces levels of bacterial populations without completely eradicating them, Bd. exovorus is believed to have some regulatory function within its ecosystem, keeping other bacterial growth under control.
Attempts have been made to introduce Bd. exovorus into living organisms as well to control the growth of potential pathogens, although they have been typically unable to establish a lasting place in an organism's microbiome without a significant amount of prey bacteria.
Pathology: How does the microbe cause disease as it interacts with the host? Describe any specific toxins or pathways that are used for invading and causing disease in the host. What treatment is used to inhibit or kill the microbe?
Pathology:
Like other members of the genus bdellovibrio, the species Bd. exovorus is not pathogenic to humans, and only predates on specific bacterial cells. It is incapable of attacking eukaryotic cells, as well as gram-positive bacterial cells. It is, however, resistant to many commonly-used antibiotics, including ampicillin, chloramphenicol, ciprofloxacin, and kanamycin.
Current Research
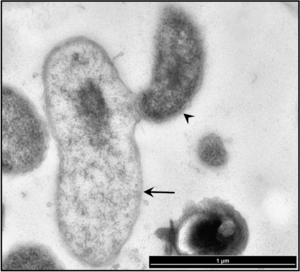
Describe recent research and findings that have been done with this organism. The research can be clinical, applied or basic research. This section should be based on 2 recent papers (10 years or less) and summarized in your own words.
Currently, Bd. exovorus strain FFRS-5 (the first strain known to attack a bacterium other than C. crescentus) is being studied as an effective treatment for the newly-emerging opportunistic pathogen Stentrophomonas maltophilia, which although it increases plant growth and in the root nodules of other plants, and kills plant pathogens, it is inherently very resistant to antibiotics, heavy metals, and polycyclic aromatic hydrocarbons. Because of this, hospital patients with compromised immune systems are susceptible to infection from S. maltophilia. Symptoms of infection include pneumonia, bacteremia, urinary tract and soft tissue infections, eye infections, endocarditis, and meningitis. The bacterium is also being increasingly found alongside P. aeruginosa in cases of cystic fibrosis. While it is not very virulent, S. maltophilia can colonize rapidly, forming biofilms in harsh environments and thus causing infections.
In order to treat infection, neither antibiotics nor phage therapy have proven effective (phages are too strain-specific and potentially transfer resistance genes to their host), leaving bacterial predators as a potential method of treatment. Because they are unable to attack eukaryotic cells, and have surprisingly high antibiotic levels themselves, they make an excellent candidate for use as a live antimicrobial. In combination with the antibiotic ciprofloxacin, Bd. exovorus was able to significantly lower biofilm masses produced by S. maltophilia in both laboratory and clinical settings as recently as 2016.
It is suggested that future studies could use a mixture of a number of Bdellovibrio and like organisms to attack and reduce infections caused by many other drug-resistant bacteria like P. aeruginosa, B. cenocepacia, E. coli, and species of Acinetobacter. This could be used to keep open wounds free of infection, and prevent the growth of biofilms on catheters and stents. Perhaps it could even be used in aerosol form to treat biofilms of pathogenic bacteria in patients with upper respiratory infections and cystic fibrosis!
Another possible future study involves "tricking" Bd. exovorus into attacking gram-positive bacteria as well. Bd. exovorus has already been observed reversibly attaching to gram-positive cell walls; it could be possible to manipulate the surface receptors to send the intermediary signals necessary to begin the growth phase.
References
[1] Koval SF, Hynes SH, Flannagan RS, Pasternak Z, Davidov Y, Jurkevitch E. Bdellovibrio exovorus sp. nov., a novel predator of Caulobacter crescentus. Int. J. Syst. Evol. Microbiol [Internet]. 2013 [cited 13 Feb 2017];(63):146–151.
[2] Bdellovibrio exovorus JSS [internet]. University of California- Integrated Microbial Genomes & Microbial Samples. [cited 2017 Feb 13] Availible from https://img.jgi.doe.gov/cgi-bin/m/main.cgi?section=TaxonDetail&page=taxonDetail&taxon_oid=2541047022
[3] Brenner DJ. 2005. Bergey's manual of systematic bacteriology. Vol. 2. The proteobacteria. Part C. The alpha-, beta-, delta-, and epsilonproteobacteria. Staley JT, editor. New York (NY): Springer. p.1040-1053.
[4] Chanyi RM, Koval SF, and Brooke JS. Stenotrophomonas maltophilia biofilm reduction by Bdellovibrio exovorus. Environmental Microbiology Reports [Internet]. 2016 [cited 31 Mar 2017]; 8:343–351.
[5] Chanyi RM. 2014. 2285: Cell Biology of the Entry of Bdellovibrio and Like Organisms [dissertation]. [London (ON)]: University of Western Ontario.
[6] Pasternak Z, Njagi M, Shani Y, Chanyi R, Rotem O, Lurie-Weinberger MN, Koval S, Pietrokovski S, Gophna U, Jurkevitch E. In and out: an analysis of epibiotic vs periplasmic bacterial predators. International Society for Microbial Ecology [Internet]. 2014 [cited 2 Apr 2017]; 8:625-635
Authored by Kip Pierce, a student of CJ Funk at John Brown University