Polysphondylium pallidum: Difference between revisions
| Line 78: | Line 78: | ||
# [https://www.ncbi.nlm.nih.gov/pubmed/21757610 Heidel AJ, Lawal HM, Felder M, Schilde C, Helps NR, et al. 2011. Phylogeny-wide analysis of social amoeba genomes highlights ancient origins for complex intercellular communication. Genome Research 21:1882-1891] | # [https://www.ncbi.nlm.nih.gov/pubmed/21757610 Heidel AJ, Lawal HM, Felder M, Schilde C, Helps NR, et al. 2011. Phylogeny-wide analysis of social amoeba genomes highlights ancient origins for complex intercellular communication. Genome Research 21:1882-1891] | ||
# [http://www.ebi.ac.uk/ena/data/view/CP001838 European Nucleotide Archive] | # [http://www.ebi.ac.uk/ena/data/view/CP001838 European Nucleotide Archive] | ||
# | # [http://download.springer.com/static/pdf/522/art%253A10.1186%252F1471-2148-11-31.pdf?originUrl=http%3A%2F%2Fbmcevolbiol.biomedcentral.com%2Farticle%2F10.1186%2F1471-2148-11-31&token2=exp=1493132809~acl=%2Fstatic%2Fpdf%2F522%2Fart%25253A10.1186%25252F1471-2148-11-31.pdf*~hmac=d37c030616e9f72978d3924bffe1f86afee4e3b46766f96e104d8937421990e1 SE Kalla, DC Queller, A Lasagni, JE Strassman. 2011. Kin discrimination and possible cryptic species in the social amoeba Polysphondylium violaceum. BMC Evolutionary Biology 11:1-11] | ||
# | # [https://books.google.com/books?id=09j_AwAAQBAJ&pg=PA46&lpg=PA46&dq=p+pallidum+food+sources&source=bl&ots=1CRpXS7PJs&sig=KtySG87nyNawP8vmrgv9IEyrw04&hl=en&sa=X&ved=0ahUKEwjr7OOC17_TAhWk1IMKHWTfAbQQ6AEIJDAA#v=onepage&q=p%20pallidum%20food%20sources&f=false Kenneth Bryan Raper: The Dictyostelids] | ||
# | # [http://tolweb.org/notes/?note_id=51 Tree of Life - Amoebae: Protists Which Move and Feed Using Pseudopodia] | ||
# | # | ||
# [http://www.newworldencyclopedia.org/entry/Slime_mold New World Encyclopedia] | # [http://www.newworldencyclopedia.org/entry/Slime_mold New World Encyclopedia] | ||
Revision as of 15:05, 25 April 2017
Classification
Domain: Eukarya
Unranked "Supergroup": Amoebozoa
Intraphylum: Mycetozoa
Class: Dictyostelea
Order: Dictyosteliida
Family: Dictyosteliidae
Genus: Polysphondylium
Species; P. palladium
Species
|
NCBI Taxonomy: Polysphondylium pallidum |
Description and Significance
Description:
Polysphondylium pallidum was first described by Edgar W. Olive in 1901 as growing on the dung of a donkey, muskrat, and rabbit in Liberia [2,4].
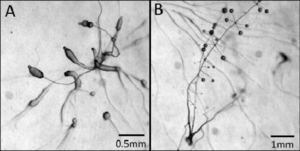
P. pallidum is a cellular slime mold of the phylum Mycetozoa. It begins its life as a single-celled amoeboid protist and lives in feces, soil, and other organic matter [1]. Like other cellular slime molds, P. pallidum can reproduce both sexually and asexually depending on its environmental conditions [1] and has two distinct stages of its life cycle: a sexual vegetative non-social stage and an asexual aggregate phase which is cued by starvation [5].
Significance:
Biologists have been particularly interested in cellular slime molds because their asexual cycles model cell differentiation in eukaryotes [2,3]. With their simple and easy-to-manipulate systems, cellular slime molds provide insight into the formation of multicellular organisms [2]. P. pallidum in particular has been used a variety of experiments because of its high levels of germination compared to other sexually reproducing slime molds, making it useful for: identifying and studying proteins involved in cellular adhesion [5]; studying the synthesis of cyclic AMP [6]; and studying the development and adaptive value of seemingly altruistic behavior of stalk cells [22].
Genome Structure
P. pallidum is predicted to contain seven linear chromosomes during the sexual, haploid stage of its lifecycle. The total genome size of these seven chromosomes is about 33Mbp, representing 12.3k coding sequences with an average gene length of 1.6kb, 32% G/C content, and a telomeric repeat sequence of TAAGGG [7]. P. pallidum is also predicted to contain a 48kb mitochrondrial genome [7]. Additionally the PPN500 strain of P. pallidum, which is commonly used in research, contains a 27kb circular plasmid with 11 predicted coding sequences [8].
P. pallidum is predicted to have diverged alongside Dictyostelium discoideum and Dictyostelium fasciculatum from a recent common ancestor about 0.6 billion years ago [7]. The P. pallidum genome contains comparably fewer transposable elements and shows high conservation of genes related primary metabolism, cytoskeletal function, and signal transduction compared to other Dictyosteliidae. Notable proteins that have been predicted in P. pallidum include polyketide synthases, which can be used to produce antibiotics and fungicides from secondary metabolites; calcium-dependent cell-adhesion proteins, which allow for extracellular cell adhesion; LagBC homologs, which are used for kin discrimination in D. discoideum; and cAMP/cGMP adenylyl cyclases ACA, ACB, ACG, sGC, and GCA [7]. cAMP and cGMP in Dictyosteliidae are secreted as signalling molecules during chemotactic cell aggregation.
Cell Structure, Metabolism and Life Cycle
P. pallidum and other Dictyostelids exhibit a unique, biphasic lifestyle. P. pallidum lives a single-celled, haploid, free-living amoeba that reproduces via mitosis (vegetative cycle). However, P. pallidum can differentiate into two forms: as part of a diploid macrocyst, similar to a zygote, created via meiosis (sexual cycle); or as part of a multicellular complex known as a fruiting body (social cycle) [9]. These phases of the P. pallidum lifecycle can be triggered by different environmental cues.
As a single-celled amoeba, P. pallidum is a chemoorganoheterotroph that feeds on bacteria and yeasts found in soils or decaying organic matter, such as Escherichia coli. This is achieved via phagocytosis [10,14]. P. pallidum, like most amoebas, is motile via the extension of pseudopods. This process mediated by extension and contraction of an actin-based microtubule network that forms a skeletal backbone in the cytoplasm [11]. Chemotaxis, in which the direction of movement is controlled by extracellular chemical signals, in P. pallidum and other Dictyostelids has been an ongoing area of scientific research. It has been found that cyclic adenosine monophosphate (cAMP), first isolated from the related Dictyostelid D. discoideum, is secreted extracellularly and plays a role in both chemotaxis and cell development [12,13]. cAMP falls into a class of molecules known as acrasins, which are required for Dictyostelid chemotaxis although not all Dictyostelids utilize cAMP. It is now understood to be a critical secondary messenger molecule in almost all eukaryotic organisms, although the slime molds are the only known organisms to make use of it as a extracellular signaling molecule. P. pallidum will form aggregates known as ‘slugs’ or ‘grexes.’ The development of these aggregates is mediated by cAMP, where P. pallidum amoebae follow the cAMP trail left by a sister cell, amplifying the signal along the way [15,16].
In response to starvation stress, P. pallidum slugs will aggregate into a single, multicellular cell mass. This mass then undergoes differential gene expression, creating a structure known as a fruiting body. The fruiting body has distinct regions, including a stalk, encystated spore head, and multiple branching whorls. The fruiting body extends vertically to a height of several millimeters, which is thought to aid in aerial and spatial dispersion of encystated spores. Rigidity of the stalk is provided both by the secretion of cellulose, and cells making up the stalk die often during the process of stalk formation [17]. Interestingly, fruiting body constituent cells are not clonal, though they do show kin discrimination when in mixed populations--ie. fruiting bodies will only form with cells of the same species [9].
Ecology and Pathogenesis
Ecology:
Polysphondylium pallidum, like other cellular slime molds, can be found world-wide living in soil, dung, leaf litter, and other decaying organic matter often found in forests and caves [1,20,21]. It spends the majority of its life feeding on bacteria as an independent amoeboid unicellular protist, acting in forest soils to maintain the balance of microbial flora [19]. In the absence of sufficient bacterial food, a starvation-induced chemical signal is emitted, causing the the individual cells to aggregate [18]. The assemblage of cells act as a single multicellular mass of approximately 100,000 cells which can move to an appropriate location and create a fruiting body [18].
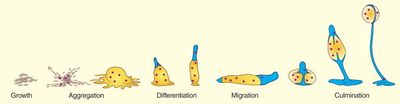
Cellular slime molds are largely considered an interesting example of seemingly altruistic behavior. As the individual cells aggregate to form a slug and subsequently a fruiting body, the cells that form the “stalk” die in the process. As an aggregate of individual cells rather than a mass of asexual clones, the cells that die to form the stalk lose their chance to reproduce and pass on their genes, leaving that benefit to only the cells forming the fruiting body [23].
Interestingly, Polysphondylium pallidum was the first recorded cellular slime mold seen to kill other slime molds [20]. The unique P. pallidum strain CK-8 isolated from Japanese forest soils was found to kill all nearby strains of Dictyostelium and Polysphondylium except for a single resistant strain (CK-9) isolated from soils very near to CK-8, found to be the same mating type as strain CK-8. As such, the CK-8 strain of P. pallidum may have a competitive advantage in nature with the ability to outcompete and kill other slime molds of differing mating types [20].
Pathogenesis:
At the present, P. pallidum has not been found to cause any diseases in humans or any other organisms.
References
- Encyclopedia of Life: Polysphondylium pallidum
- University of Califronia: Introduction to Slime Molds
- Mirfakhrai, M., Tanaka, Y., & Yanagisawa, K. (1990). Evidence for mitochondrial DNA polymorphism and uniparental inheritance in the cellular slime mold Polysphondylium pallidum: effect of intraspecies mating on mitochondrial DNA transmission. Genetics, 124(3), 607-613.
- Olive, Edgar W. (1901). "A preliminary enumeration of the Sorophoreae". Proceedings of the American Academy of Arts and Sciences 37 (12): 333–344. doi:10.2307/20021671.
- Rosen, S. D., Simpson, D. L., Rose, J. E., & Barondes, S. H. (1974). Carbohydrate-binding protein from Polysphondylium pallidum implicated in intercellular adhesion. Nature, 252(5479), 128-151.
- Konijn, T. M., CHANG, Y. Y., & BONNER, J. T. (1969). Synthesis of cyclic AMP in Dictyostelium discoideum and Polysphondylium pallidum. Nature, 224(5225), 1211-1212.
- Heidel AJ, Lawal HM, Felder M, Schilde C, Helps NR, et al. 2011. Phylogeny-wide analysis of social amoeba genomes highlights ancient origins for complex intercellular communication. Genome Research 21:1882-1891
- European Nucleotide Archive
- SE Kalla, DC Queller, A Lasagni, JE Strassman. 2011. Kin discrimination and possible cryptic species in the social amoeba Polysphondylium violaceum. BMC Evolutionary Biology 11:1-11
- Kenneth Bryan Raper: The Dictyostelids
- Tree of Life - Amoebae: Protists Which Move and Feed Using Pseudopodia
- New World Encyclopedia
- Dela Cruz, T. E. E., Santiago, K. A. A., Ramirez, C. S. P., Torres, J. M. O., Dagamac, N. H. A., Yap, J., ... & Yulo, P. R. J. (2011). Occurrence of Cellular Slime Molds (Dictyostelids) in Subic Bay Natural Forest Reserve, Zambales, Philippines. Philippine Journal of Systematic Biology, 5, 99-104.
- Mizutani, A., Hagiwara, H., & Yanagisawa, K. (1990). A killer factor produced by the cellular slime mold Polysphondylium pallidum. Archives of microbiology, 153(5), 413-416.
- Landolt, J. C., Stephenson, S. L., & Slay, M. E. (2006). Dictyostelid cellular slime molds from caves. J Cave Karst Stud, 68, 22-26.
- Bonner, J. T. (1982). Evolutionary strategies and developmental constraints in the cellular slime molds. The American Naturalist, 119(4), 530-552.
- Shaulsky, G., & Kessin, R. H. (2007). The cold war of the social amoebae. Current Biology, 17(16), R684-R692.
Author
Page authored by Benjamin Braude and Alexandra Canzoneri, students of Prof. Jay Lennon at Indiana University.
