Bacillus Subtilis Soil Project: Difference between revisions
No edit summary |
|||
| Line 11: | Line 11: | ||
Family: Bacillaceae | Family: Bacillaceae | ||
[[File:IMG_2840(1).jpg]] | |||
===Species=== | ===Species=== | ||
Revision as of 18:18, 4 May 2018
Classification
Domain: Bacteria
Phylum: Firmicutes
Class: Bacilli
Order: Bacillales
Family: Bacillaceae
Species
|
NCBI: Taxonomy |
Bacillus subtilis
Habitat Information
Description of location and conditions under which the organism was isolated:
Date: 1.25.2018
Temperature: 58° F
Recent rainfall: 0 inches
Depth: Surface to 2”
Grid coordinates: 30.20144°, -97.88822°
Soil type number and name from NRCS soil map:
Name: Volente silty clay loom, 1 to 8 percent slopes
Unit key and symbol: 39325833, f66r
Description: The location the organism was isolated was a grassy field between a soccer field, parking lot, and children’s playground. There was full sun, little traffic near the area, and used often by local residents from the suburban area.
Description and Significance
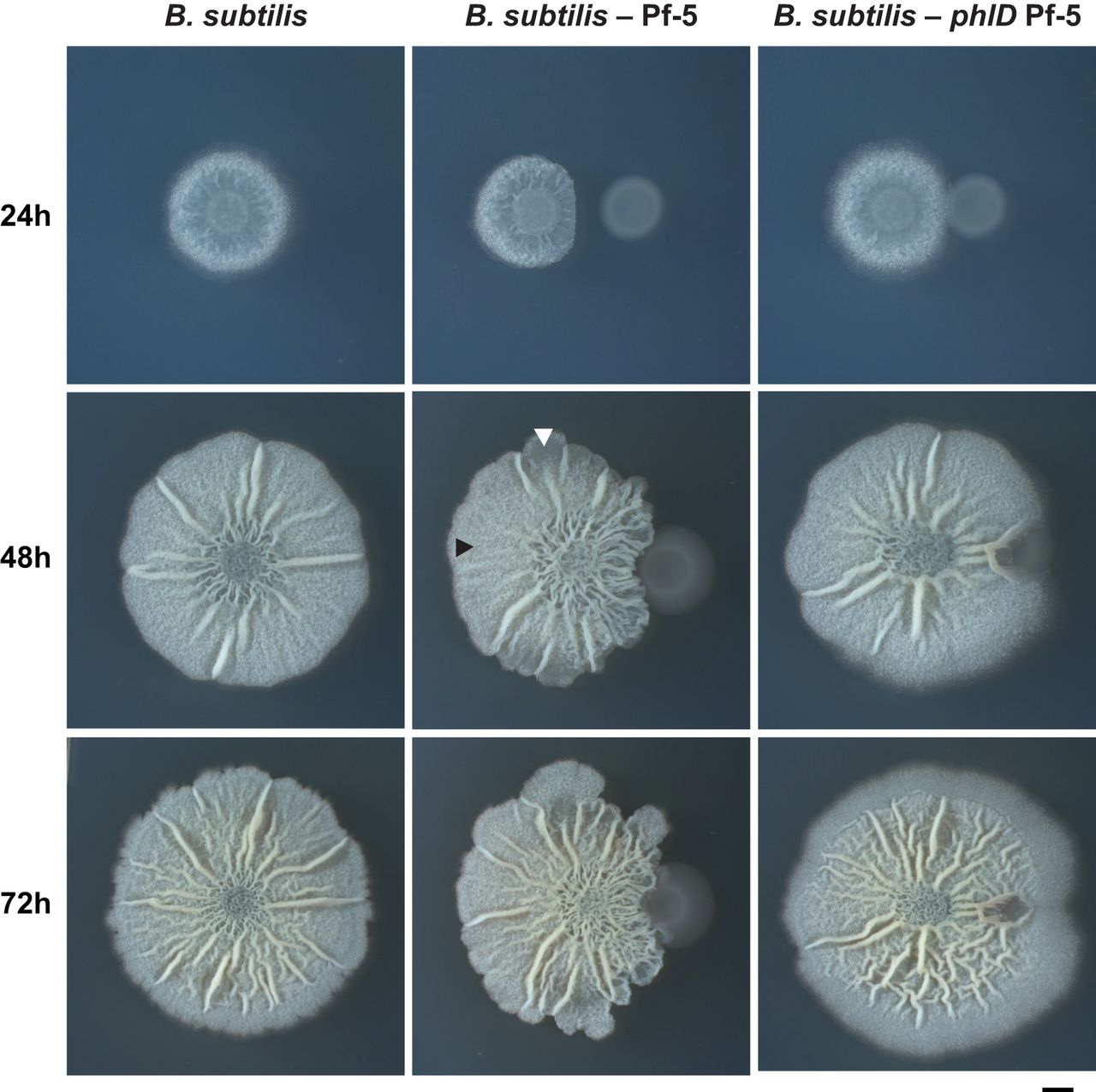
The appearance of a colony of B. Subtilus depends of the amount of time it is allowed to grow. At around 24 hours of incubation, the colony’s appearance is a white convex, circle with smooth edges. In general, if allowed to grow, the colony’s appearance at 48 hours would be whitish in color, with irregular margins and a rough, wrinkled, textured surface with ridges and furrows. At 72 hours and beyond, the colony continues to grow, but remains the same coloration with irregular margins and rough texture as seen in 48 hours of growth. (1)
Via microscopy, individual cells are colorless rod shaped bacilli, readily taking up staining via various methods. Depending on environmental conditions, samples may include bacteria in various stages of formation of an endospore. (2) (3)
An interesting fact about Bacillus subtilis is that there are strains that have been identified for production of Bacteriocin (4) and other antimicrobial compounds(5).
Genome Structure
The organism has 4,214,810 base pairs which codes for 4100 protein coding genes. (6) It has a single, circular chromosome.(7)(8)
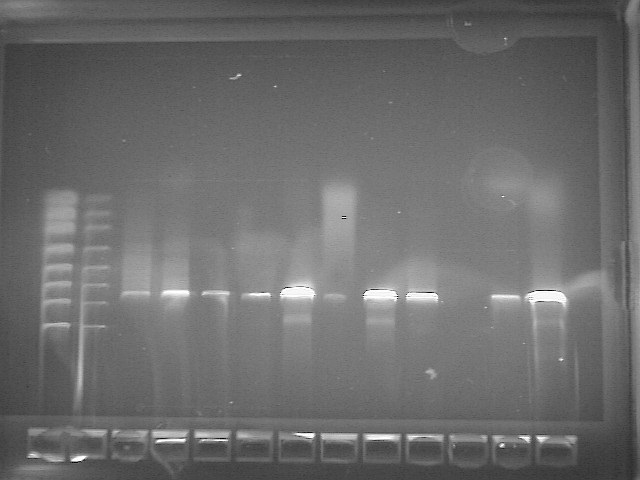
The PCR imaging results for our class is below. Our bacteria's DNA was run in well #5.
HMN1-Forward_A06.ab1 937 letters, trimmed about 40 b/p (9)
TTGNNGCGTANGGGCTCGCAGGCGGTTTCTTAAGTCTGATGTGAAAGCCCCCGGCTCAACCGGGGAGGGTCATTGGAAACTGGGGAACTTGAGTGCAGAAGAGGAGAGTGGAAT TCCACGTGTAGCGGTGAAATGCGTAGAGATGTGGAGGAACACCAGTGGCGAAGGCGACTCTCTGGTCTGTAACTGACGCTGAGGAGCGAAAGCGTGGGGAGCGAACAGGATTAG ATACCCTGGTAGTCCACGCCGTAAACGATGAGTGCTAAGTGTTAGGGGGTTTCCGCCCCTTAGTGCTGCAGCTAACGCATTAAGCACTCCGCCTGGGGAGTACGGTCGCAAGAC TGAAACTCAAAGGAATTGACGGGGGCCCGCACAAGCGGTGGAGCATGTGGTTTAATTCGAAGCAACGCGAAGAACCTTACCAGGTCTTGACATCCTCTGACAATCCTAGAGATA GGACGTCCCCTTCGGGGGCAGAGTGACAGGTGGTGCATGGTTGTCGTCAGCTCGTGTCGTGAGATGTTGGGTTAAGTCCCGCAACGAGCGCAACCCTTGATCTTAGTTGCCAGC ATTCAGTTGGGCACTCTAAGGTGACTGCCGGTGACAAACCGGAGGAAGGTGGGGATGACGTCAAATCATCATGCCCCTTATGACCTGGGCTACACACGTGCTACAATGGACAGA ACAAAGGGCAGCGAAACCGCGAGGTTAAGCCAATCCCACAAATCTGTTCTCAGTTCGGATCGCAGTCTGCAACTCGACTGCGTGAAGCTGGAATCGCTAGTAATCGCGGATCAG CATGCCGCGGTGAATACGTTCCCGGGCCTTGTACACACCGCCCGTCACACCACGAGAGTTTGTAACACCCGAAGTCGGTGAGGTAACCTTTTAGGAGCCAGCCGCCGAANGTGG GACAGATGATTGGGGTGAANTCGTA
HMN2-Reverse_B06.ab1 936 letters, trimmed about 20 b/p (10) TCGGNGGNTGGCTCCTAAAAGGTTACCTCACCGACTTCGGGTGTTACAAACTCTCGTGGTGTGACGGGCGGTGTGTACAAGGCCCGGGAACGTATTCACCGCGGCATGCTGATC CGCGATTACTAGCGATTCCAGCTTCACGCAGTCGAGTTGCAGACTGCGATCCGAACTGAGAACAGATTTGTGGGATTGGCTTAACCTCGCGGTTTCGCTGCCCTTTGTTCTGTC CATTGTAGCACGTGTGTAGCCCAGGTCATAAGGGGCATGATGATTTGACGTCATCCCCACCTTCCTCCGGTTTGTCACCGGCAGTCACCTTAGAGTGCCCAACTGAATGCTGGC AACTAAGATCAAGGGTTGCGCTCGTTGCGGGACTTAACCCAACATCTCACGACACGAGCTGACGACAACCATGCACCACCTGTCACTCTGCCCCCGAAGGGGACGTCCTATCTC TAGGATTGTCAGAGGATGTCAAGACCTGGTAAGGTTCTTCGCGTTGCTTCGAATTAAACCACATGCTCCACCGCTTGTGCGGGCCCCCGTCAATTCCTTTGAGTTTCAGTCTTG CGACCGTACTCCCCAGGCGGAGTGCTTAATGCGTTAGCTGCAGCACTAAGGGGCGGAAACCCCCTAACACTTAGCACTCATCGTTTACGGCGTGGACTACCAGGGTATCTAAT
Cell Structure, Metabolism and Life Cycle
Interesting features of cell structure; how it gains energy; what important molecules it produces. B. subtilis is a rod-shaped bacterium arranged in either single cells, small clumps, or short chains. Its cell wall consists of a thick peptidoglycan layer. B. subtilis is apart of the kingdom Bacteria, which means this organism has a single circular chromosome within the nucleoid region of its cytoplasm. B. subtilis has a helical cytoskeleton composed of a single protein. (11)
B. subtilis is a motile organism through use of its flagella, which is a whip-like appendage used for movement. Specifically, B. subtilis has peritrichous flagella, meaning has flagella projecting in all directions around the cell. (11)
B. subtilis exhibits endospore formation. Endospores are dormant durable structures often created from a vegetative cell in response to nutrient deprivation are produced through the process sporulation. During this process, a thick layer of peptidoglycan and spore coat form around a copy of the cell’s DNA and part of the cytoplasm. This allows the bacteria to survive under harsh conditions such as high temperatures, chemical damage, etc. This is so the chromosome can be protected within and then, and the bacteria genetic material is not harmed. (11) Another important note of B. subtilis producing endospores is this means it cannot readily be killed by many antimicrobial treatments. (12)
In terms of obtaining energy, Bacillus subtilis is classified as facultative anaerobe, meaning can live with or without oxygen. This bacterium can produce ATP through nitrate ammonification or fermentation. During nitrate ammonification, nitrate is eventually reduced to ammonia by the respiratory nitrite reductase. This bacterium can use nitrite or nitrate to be used as a terminal electron acceptor. During anaerobic fermentation, carbon sources are transformed by pyruvate and end products include lactate, acetoin, 2,3-butanediol, ethanol, acetate, and succinate. NAD+ regeneration utilizes the enzyme lactate dehydrogenase and this enzyme also converts pyruvate into lactate. These processes produce different ATP yields, and B. subtilis compensates for this imbalance by using a specific regulatory system that allows for the most efficient ATP production. According to research, not all of the parts of this regulatory system are known. (13)
Industrial sectors often use B. subtilis because of the production of important production of hyaluronic acids, specifically proteases. (13) Proteases are enzymes frequently used in detergents, pharmaceuticals, food and agricultures industries around the world. New technology is even being created in order to meet the demand for this protease-producing bacterium. (12) Also according to studies, B. subtilis is free of endotoxins and exotoxins, which generally recognizes it as safe (GRAS). (14) This makes B. subtilis more favorable in being used in food production over some gram-negative bacteria’s. (14)
Physiology and Pathogenesis
Biochemical characteristics, enzymes made, other characteristics that may be used to identify the organism; contributions to environment (if any). If relevant, how does this organism cause disease? Human, animal, plant hosts? Virulence factors, as well as patient symptoms.
Biochemical characteristics are as follows:
Gram Reaction: +
Capsule Stain: -
Endospore Stain: +
Motility results: +
Phenol Red Broth:
Sucrose: +
Lactose: -
Glucose: +
Starch Hydrolysis: +
Gelatin Hydrolysis: +
DNA Hydrolysis: -
Lipid Hydrolysis: +
Methyl Red: -
Voges Proskauer: +
Citrate test: +
SIM Tests (3 in 1): -
Nitrate Reduction Test: +
Urea Hydrolysis: -
Triple Sugar Iron Agar: K/A, G
Oxidase: -
Eosin Methylene Blue Agar: +
Hektoen Enteric Agar: -
MacConkey Agar: -
Decarboxylation of Arginine, Lysine, Orthinine: +
Phenylalanine Deaminase: +
Catalase: +
Bile Esculin: +
Blood Agar: alpha hemolysis
Mannitol Salt Agar: +
Phenylethyl Alcohol Agar: +
Bacillus Subtilis is a gram positive, rod shaped organism that can be found growing in soil as well as the gastrointestinal tract of humans. The microrganisim B. Subtilis tested positive for catalase, lipase, and amylase. Uses citrate as its sole carbon source also positive for carbohydrate fermentation. B. Subtilis is considered non-pathogenic or toxic and does not cause any disease, it can be found in the gastrointestinal tract of humans but this is very rare. Many studies have been conducted by the FDA and each concluded that Bacillus Subtilis and other microorganism derived are considered safe.
References
Description and Significance section:
1. Image Credit: American Society for Microbiology, Journal of Bacteriology. vol. 197 no. 13 2129-2138 (2015, July) http://jb.asm.org/content/197/13/2129/F3.expansion.html
2. Earl AM, Losick R, Kolter R. Ecology and genomics of Bacillus subtilis. Trends in microbiology. 2008;16(6):269. doi:10.1016/j.tim.2008.03.004.
3. Image Credit: "File:Bacillus subtilis Spore.jpg." Wikimedia Commons, the free media repository. 23 Oct 2013, 06:01 UTC. 4 May 2018, 16:15 https://commons.wikimedia.org/w/index.php?title=File:Bacillus_subtilis_Spore.jpg&oldid=107715257
4. Sharmila, P.S., CHARACTERIZATION AND ANTIBACTERIAL ACTIVITY OF BACTERIOCIN PRODUCING BACILLUS SUBTILIS ISOLATED FROM RAW MILK. International Journal on Applied Bioengineering,(2015, July) Vol 9, Issue 2 https://pdfs.semanticscholar.org/08a2/380533115f7aedefe9c354222c04c65df21e.pdf
5. Ouoba, L.I.I, Antimicrobial activity of Bacillus subtilis and Bacillus pumilus during the fermentation of African locust bean (Parkia biglobosa ) for Soumbala production. Journal of Applied Microbiology. (2006) https://onlinelibrary.wiley.com/doi/pdf/10.1111/j.1365-2672.2006.03156.x
Genome Structure section:
6. Kunst, F., The complete genome sequence of the gram-positive bacterium Bacillus subtilis., Nature. (1997) https://www.ncbi.nlm.nih.gov/pubmed/9384377
7. Moszer, I. The complete genome of Bacillus subtilis: from sequence annotation to data management and analysis. FEBS Letters. (1998) https://www.sciencedirect.com/science/article/pii/S0014579398006206
8. Medigue, C., Analysis of a Bacillus subtilis genome fragment using a co-operative computer system prototype. Gene. (1995) https://www.sciencedirect.com/science/article/pii/037811199500636K
9. NIH Blast Suite. https://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE_TYPE=BlastSearch
10. NIH Blast Suite. https://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE_TYPE=BlastSearch
Metabolism section: 11. Bauman, R. W., Ph.D., Crosby, C. D., Ph.D., FNP-C, PA-C, Fulks, J., Ed.D., Lammert, J. M., Ph.D., Machunis-Masuoka, E., Ph.D., & Montgomery, J. E., MSN, RN. (2015,2012,2009). Microbiology With Disease By Body System (4th ed.). Pearson Education, Inc.
12. Westers, L., Westers, H., & Quax, W. (2004, May 17). Bacillus subtilis as cell factory for pharmaceutical proteins: A biotechnological approach to optimize the host organism. Retrieved from https://www.sciencedirect.com/science/article/pii/S0167488904000837
13. Marino, M., Ramos, H. C., Hoffmann, T., Glaser, P., & Jahn, D. (2001). Modulation of Anaerobic Energy Metabolism of Bacillus subtilis by arfM (ywiD). Journal of Bacteriology, 183(23), 6815–6821. http://doi.org/10.1128/JB.183.23.6815-6821.2001
14. Widner, B., Behr, R., Von Dollen, S., Tang, M., Heu, T., Sloma, A., … Brown, S. (2005). Hyaluronic Acid Production in Bacillus subtilis. Applied and Environmental Microbiology, 71(7), 3747–3752. http://doi.org/10.1128/AEM.71.7.3747-3752.2005
Author
Page authored by Nathan Zuck, Melissa Bradley, and Hailey Langston, students of Prof. Kristine Hollingsworth at Austin Community College.