|
|
| Line 34: |
Line 34: |
| ==Genome Structure== | | ==Genome Structure== |
|
| |
|
| <br /> The genome of the circular chromosome of the PCC 7120 strain of ''Anabaena'' has been sequenced. It contains 1 chromosome with 6,413,771 base pairs with 5,368 predicted protein-coding regions, and six plasmids ranging from 408,101 base pairs to 5,584 base pairs. The sequencing of the anabaena genome will aid researchers in studying the genetics and physiology of cellular differentiation, (as exhibited in the heterocycsts of ''Anabaena'') pattern formation, and nitrogen fixation. [http://www.kazusa.or.jp/cyano/Anabaena/index.html Distribution maps for chromosome and six plasmids, courtesy of Cyanobase.] The genome of <font size="+0">''Anabaena variabilis'' ATCC 29413</font><font size="3"> has also been completed. Unlike most other strains of ''Anabaena'', <font size="+1">''Anabaena sp. strain ATCC 29413 '' </font></font> <font size="3"> differenciates between their hetrocyst and akinetes forms. This provides a model for the study of gene differentiation in the formation of heterocysts and akinetes. Akinetes are the resting forms of a organism.</font><font size="3"> </font> | | <br /> The genome of the circular chromosome of the PCC 7120 strain of ''Anabaena'' has been sequenced. It contains 1 chromosome with 6,413,771 base pairs with 5,368 predicted protein-coding regions, and six plasmids ranging from 408,101 base pairs to 5,584 base pairs. The sequencing of the anabaena genome will aid researchers in studying the genetics and physiology of cellular differentiation, (as exhibited in the heterocycsts of ''Anabaena'') pattern formation, and nitrogen fixation. [http://www.kazusa.or.jp/cyano/Anabaena/index.html Distribution maps for chromosome and six plasmids, courtesy of Cyanobase.] The genome of ''Anabaena variabilis'' ATCC 29413 has also been completed. Unlike most other strains of ''Anabaena'', ''Anabaena sp. strain ATCC 29413'' differenciates between their hetrocyst and akinetes forms. This provides a model for the study of gene differentiation in the formation of heterocysts and akinetes. Akinetes are the resting forms of a organism. |
|
| |
|
| ==Cell Structure and Metabolism== | | ==Cell Structure and Metabolism== |
Revision as of 18:46, 31 May 2006
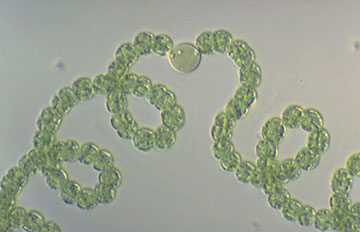
Anabaena flos-aquae f. flos-aquae. From the Microbial Culture Collection

Anabaena oumiana. From the Microbial Culture Collection
Classification
Higher order taxa:
Bacteria; Cyanobacteria; Nostocales; Nostocaceae; Anabaena
Species:
Anabaena sp. PCC 7120, Anabaena flos-aque, Anabaena aequalis
Description and Significance
Anabaena provides a model for the study of gene differentiation in the formation of heterocysts. The recent sequencing of the genome of Anabaena sp. strain PCC 7120 is yet another step towards understanding this process.
Genome Structure
The genome of the circular chromosome of the PCC 7120 strain of Anabaena has been sequenced. It contains 1 chromosome with 6,413,771 base pairs with 5,368 predicted protein-coding regions, and six plasmids ranging from 408,101 base pairs to 5,584 base pairs. The sequencing of the anabaena genome will aid researchers in studying the genetics and physiology of cellular differentiation, (as exhibited in the heterocycsts of Anabaena) pattern formation, and nitrogen fixation. Distribution maps for chromosome and six plasmids, courtesy of Cyanobase. The genome of Anabaena variabilis ATCC 29413 has also been completed. Unlike most other strains of Anabaena, Anabaena sp. strain ATCC 29413 differenciates between their hetrocyst and akinetes forms. This provides a model for the study of gene differentiation in the formation of heterocysts and akinetes. Akinetes are the resting forms of a organism.
Cell Structure and Metabolism
|
|
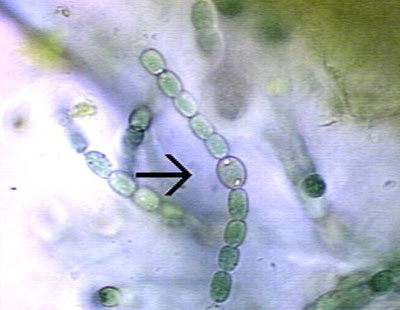
Anabaena are heterocyst-forming, photoautotrophic cyanobacteria that perform oxygenic photosynthesis. Anabaena grow in long filaments of vegetative cells. During times of low environmental nitrogen, about one cell out of every ten will differentiate into a heterocyst. Heterocysts then supply neighboring cells with fixed nitrogen in return for the products of photosynthesis, that they can no longer perform. This separation of functions is essential because the nitrogen fixing enzyme in heterocysts, nitrogenase, is unstable in the presence of oxygen.
Due to the necessity of keeping nitrogenase isolated from oxygen, heterocysts have developed elements to maintain a low level of oxygen within the cell. To prevent the entrance of oxygen into the cell, the developing heterocyst builds three additional layers outside the cell wall, giving it its characteristic enlarged and rounded appearance, thus the rate of oxygen diffusion into heterocysts is 100 times lower than of vegetative cells. One layer creates an envelope polysaccharide layer where the nitrogen is fixed in a oxygen-restricted milieu. To lower the amount of oxygen within the cell, the presence of photosystem II is eliminated, and the rate of respiration is stepped up to use up excess oxygen.
|
Ecology
It is commonly accepted that cyanobacteria played a key role in the manufacturing of oxygen during the Precambrian period of earth's history. They perform oxygenic photosynthesis, very similar to that of eukaryotic plants and algae, by utilizing water as a reductant source, consequently producing molecular oxygen. Heterocysts, mentioned above, are terminally differentiated cells that specialize in nitrogen fixation. It is believed that the capacity for developing heterocysts first developed when the concentration of oxygen in the atmosphere (largely produced by cyanobacteria) reached levels high enough to cause the inactivation of the nitrogen-fixing enzyme nitrogenase.
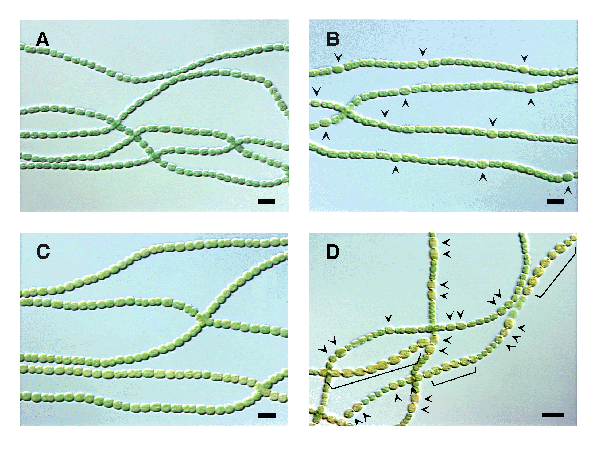
The mechanism of cellular differentiation, or the gene that initiates heterocyst formation has been the focus of much research. To date researchers have found two genes that control both the initiation of heterocyst formation and the frequency of differentiated cells. The product of the hetR gene is required for heterocyst differentiation and is only transcribed in differentiating cells. The patS gene regulates the frequency at which cells develop into heterocysts, or essentially inhibits heterocyst formation. The two genes work very differently as evidenced when studying cells carrying mutant genotypes. Cells with mutant hetR genes do not differentiate at all, where as cells with mutant patS genes differentiate too often. Conversely, cells that carry extra copies of the hetR gene differentiate additional heterocysts, and cells with extra copies of the patS gene do not differentiate at all. {| width="100%"
|
|
Figure A shows normal Anabaena grown in a complete medium. Figure B shows normal Anabaena growing in a medium lacking nitrogen, arrows indicate heterocysts. Figure C shows an overexpression of the patS gene, which inhibits heterocyst formation even in nitrogen-depleted conditions. Figure D shows the deletion of the patS gene resulting in the differentiation of additional heterocysts, arrows and brackets indicate heterocysts. From Texas A&M University
|
References
Updated May 31, 2006
Cyanobase: Anabaena sp. PCC 7120
ExPaSy Molecular Biology Server: Heterocyst or not Heterocyst?
Guocun Huang, Qing Fan, Sigal Lechno-Yossef, Elizabeth Wojciuch, C. Peter Wolk, Takakazu Kaneko, and Satoshi Tabata. Clustered Genes Required for the Synthesis of Heterocyst Envelope Polysaccharide in Anabaena sp. Strain PCC 7120. J Bacteriol. 2005 February; 187(3): 1114-1123.
Horne, A. J., J. E. Dillard, D. K. Fujita, C. R. Goldman. Nitrogen Fixation in Clear Lake California. II. Synoptic Studies on the Anabaena Bloom. American Society of Limnology and Oceanography. 1972, Vol. 17, Issue 5, Pg 702 (10 of 11).
Kenyon College: Heterocyst formation in Anabaena and other cyanobacteria
Microbiology and Molecular Biology Reviews: Nitrogenase
Masayuki, Ohmori et al. 2001. Characterization of genes encoding multi-domain proteins in the genome of the filamentous nitrogen-fixing cyanobacterium Anabaena sp. strain PCC 7120. DNA Research, 8: 271-284.
Takakazu, Kaneko et al. 2001. Complete genomic sequence ot the filamentous nitrogen-fixing cyanobacterium Anabaena sp. strain PCC 7120. DNA Research, 8: 205-213.
University of Chicago: How Cyanobacteria Count to 10