The Human Gut Microbiome: Difference between revisions
| Line 1: | Line 1: | ||
==Introduction== | ==Introduction== | ||
[[Image:humandigestion.jpg|thumb|200px|right|Figure 1: This illustration depicts the human digestive system. Microbes live throughout the digestive system to help metabolize food. The photo credit for this image belongs the National Institute of Diabetes and Digestive and Kidney Diseases [https://www.niddk.nih.gov/news/media-library/18162/ NIH].]] | [[Image:humandigestion.jpg|thumb|200px|right|Figure 1: This illustration depicts the human digestive system. Microbes live throughout the digestive system to help metabolize food. The photo credit for this image belongs the National Institute of Diabetes and Digestive and Kidney Diseases [https://www.niddk.nih.gov/news/media-library/18162/ NIH].]] | ||
Revision as of 03:31, 28 April 2023
Introduction
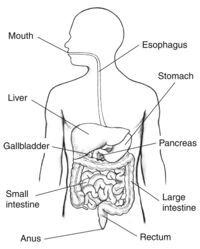
Nutrients from your food are absorbed into your body through complex pathways that occur in the digestive system. Humans need nutrients like proteins, fats, carbohydrates, vitamins, and water in order to survive which are obtained via food digestion. Much of these nutrients are absorbed into the blood through enzymatic pathways. However, some food components are unable to be digested by human enzymes, which is where microbes come into play. Some microbes in the digestive tract, which usually live along the epithelial cells which make up the lining of the digestive tract, are able to digest certain food components that humans are not. Other microbes in the intestines live without a specified benefit to the host and some other microbes can be detrimental to the human host [1]. The digestive system is made up of the gastrointestinal tract-- also known as the GI tract-- liver, pancreas, and gall bladder. This anatomy is shown in figure 1[2]. To reach the nutrients, the body must first break down the food. The first and most simple step to this process is chewing and swallowing food which then begins its pathway through the GI tract.
After being chewed and swallowed the food travel down the esophagus, then to the stomach, then to the small intestine, large intestine, and then to the anus [3] . All the organs that make up the GI tract are hollow to allow the food to pass through. Once the food has passed through the esophagus and into the stomach it is broken down using digestive juices into a liquid form called chyme. Compared to the microbiomes of the small and large intestine, the stomach does not have a large microbiome. This is because the highly acidic environment, as well as other components created by the stomach make the stomach inhabitable for many bacteria. One notable bacterium that lives in the stomach is Helicobacter pylori (H. pylori). H. pylori is in the stomach of most humans but in some cases can cause illnesses such as gastritis or ulcers. It is unique in its ability to use enzymes to neutralize the acidic environment of the stomach.
The chyme produced by the stomach then travels through the small intestine--where most nutrients are absorbed. There are a few genera that are commonly found in the small intestine including Streptococcus and Veillonella. They are known for breaking down and metabolizing carbohydrates. They compete directly with the host for food components. These bacteria do not have a direct beneficial effect on the host, but it is possible that one bacterium, called Bacteroides thetaiotaomicron (B. thetaiotaomicron), has the ability to keep the small intestine from producing too much fucose. The microbiome of the large intesting has the largest plethora of bacteria. Once the chyme has passed into the large intestine, it is depleted of most of the nutrients and the microbes then digest what the host cannot. The large intestine introduces the chyme to a plethora of colonic bacteria. These bacteria are able to digest certain carbohydrates, particularly those from plant derived foods, that the small intestine was not able to. Vitamin B production is also a critical function of the large intestine because it sources the body with the vitamin B that is not ingested from food sources. Vitamin B also is used to produce the energy used by the large intestine. Other functions of the large intestine include, but are not limited to, water absorption via osmosis and the passing of waste to the anus.
Stomach Microbiota
The main purpose of the stomach is to further break down the chewed food. The stomach is able to do this by producing digestive juices made up of stomach acid and enzymes from glands in the stomach lining. Many studies have analyzed the digestive juices for their microbiota. A study done by Bik et al in 2005 analyzed the stomach microbiota for the first time using a small subunit 16S rDNA clone library approach and found five dominant genera including Streptococcus, Prevotella, Rothia, Fusobacterium and Veillonella. Other studies conducted in later years using the same method found other genera including Neisseriae, Haemophilus, Porphyromonas, Rothia, Propionibacterium, and Lactobacillus.
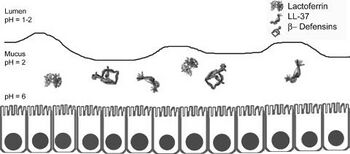
However, many of these bacteria may be transient in the stomach since the stomach is open to a swelling of bacteria from the oral cavity. For example, Lactobacilli is known to be present in the mouth for the purpose of converting the nitrate found in food and saliva into nitrite which can be converted into nitric oxide by gastric juice in the stomach to produce a useful antimicrobial agent [4] . Most microbes in general do not survive in the stomach due to its high acidity. On top of its high acidity, the stomach utilizes its gastric mucosa to produce other factors to kill bacteria that are transported from the oral cavity including peptides B-defensins 1 and 2 and LL-37 as shown in figure 2. These peptides fight off many species of bacteria. Also, an enzyme called lysozyme degrades bacterial peptidoglycan. Proteins in the stomach can also inhibit bacterial growth including lactoferrin and surfactant protein D. Lactoferrin inhibits bacterial colonization by reducing extracellular Fe3+ and surfactant protein D brings the bacteria into close proximity to each other. Finally, other mucosal components such as alpha-1,4-GluNAC-capped O-glycans, which disrupt the biosynthesis of cholesteryl-α-D-glucopyranoside, a component of the H. pylori cell wall [5] . Many bacteria that are specialized to live within the human stomach are still unknown, but among the 128 identified phylotypes identified by Bik et al in 2005, one particularly interesting sequence identified was that belonging to Deinococcus/ Thermus phylum. This phylum is known to live in extreme environments such as hot springs and radioactive sites [6].
Although much is still unknown about the stomach microbiota, one famous bacterium in the stomach is H. pylori. About 50% of the human population is known to be infected with this bacterium, but most have no symptoms. H. pylori is associated with an increased risk of duodenal ulcer disease, gastric ulcer disease, gastric adenocarcinoma, and gastric lymphoma. H. pylori is a gram-negative bacterium that elicits humoral and cellular immune responses. H. pylori is the only known bacteria to colonize in the gastric mucosa and crosstalk with the host. It does not partake in nutrient absorption but instead is the known cause of gastritis. It can survive the highly acidic environment by using its urease enzyme to produce ammonia and neutralize the acid [4] . Strains of H. pylori that can cause infection have flagella that mobilize it so that it can pass from the lumen, which has a pH between 1-2 to the mucus layer that lines the epithelial cells and has a pH of 2. It is then able to travel through the mucus layer to its ideal environment on the epithelial cell surface which has a pH of 6 [5] . The bacteria is able to attach on to the cell surface via outer membranes proteins that include BabA, SabA, AlpA, AlpB, and HopZ. This adherence allows for the activation of signaling pathways that allow for toxins to enter the cells which incurs gastric mucosal inflammation, production of autoantibodies, and parietal cell loss[5].
Small Intestine
After being liquified, the stomach contents enter the next phase of the GI tract, the small intestine. Nutritional uptake occurs mainly in the small intestine. The most prominent genera of bacteria are Streptococcus and Veillonella. Cultured isolates of the Streptococcus genus include S. mitis, S. bovis (s equnis), and S. salivaris. These bacteria metabolize sugars that come from food components in order to release immune responses in the gut. S. mitis metabolizes N-acetyl glucosamine and raffinose. S. bovis metabolizes L-Arabinose and raffinose. S. salvarius metabolizes L-Arabinose and Trehalose [7] . A study done by Bogert et al in 2013 analyzed both Streptococcus and Veillonella strains of bacteria found in the human small intestine and concluded that it is possible that Streptococcus populations play a major role in the primary carbohydrate metabolism in the small intestine [8] . Veillonella, on the other hand, ferment the lactic acid that is produced by streptococci to create acetic acid and propionic acid [7]. A 2012 study conducted by Zoetendal et al concluded that the microbiome of the small intestine is highly dependent on available carbohydrates, which supports Bogert et al’s findings that Streptococcus is responsible for carbohydrate metabolism. Zoetendal et al did a comparative functional analysis to determine that simple carbohydrate transport phosphotransferase systems, central metabolism, and biotin production are represented in the small intestine [9] . It is important for these bacteria to have strong metabolism pathways because they directly compete with the host.
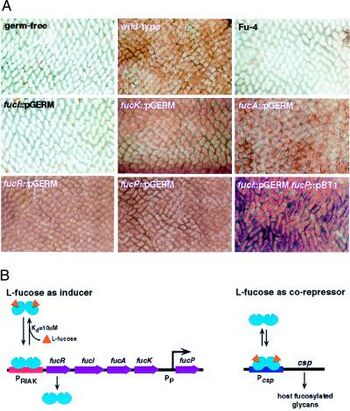
An example of a possible symbiotic relationship between the human small intestine and bacteria is that of the B. thetaiotaomicron life cycle. It has been demonstrated in mice that there is fucosylated glycan production in the distal small intestine. B. thetaiotaomicron has evolved to be able to sense the availability of carbohydrates in the surrounding environment and use fucose as an energy source [1]. It picks up fucose, which is a terminal a-linked sugar, by secreting an enzyme called a-frucosidase to obtain the monosaccharide fucose and import it as well as metabolize it. When the bacteria sense a scarcity of fructose, specifically, in the lumen it makes the gut epithelium express fucosylated glycans at a higher rate so that it can use them as an energy source . This signaling pathways is shown in figure 3. When L-fucose (fucose) is present it acts as a corepressor by binding to the control of signal production (csp) and stops the host from producing more fucose. The fucose acts through FucR, which is a repressor that is a molecular sensor of fucose availability [10] . This tightly coordinated relationship between the host and the bacteria keep the host from overproducing hydrolysable epithelial fucose. It is possible that this relationship also benefits the health of the small intestine by disallowing the colonization of pathogens that are fucosylated glycan-adhesive.
Large Intestine
One the food contents pass through the small intestine they enter the large intestine, which is approximately five feet long. At this point the food contents are mostly nutrient depleted. The large intestine has three main functions including absorbing water and electrolytes, producing and absorbing vitamins, and forming and moving waste toward the anus. Water absorption is conducted by osmosis and sodium and other elements are absorbed through biochemical channels. Motility of waste is conducted by smooth muscle with the aid of mucus layers. The production and absorption of vitamins, on the other hand, are highly dependent on bacteria. Colonic bacteria most famously produce vitamins B and K [11] . Colonic bacteria use both diet derived and host-derived food sources for energy and growth. They contain significantly more genes than humans that or able to digest food components. Most of their metabolism is done through fermentation.
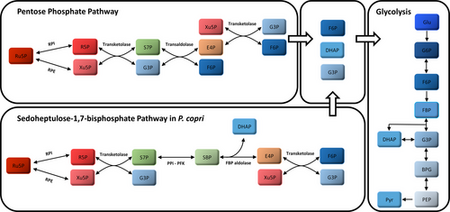
Some carbohydrates, including those in plant-derived foods, cannot be digested in the small intestine and therefore are available for microbial digestion in the large intestine. Pentose is fermented via Prevotella copri (P. copri). P. copri, which is a part of the Bacteroidetes phylum, uses plant-derived fibers to produce acetate, formate, and succinate [12]. Genes encoded in P. copri are used to uptake pentose and convert it into important intermediates including L-ribulose-5-phosphate (Ru5P) and D-xylulose-5-P (Xu5P). Ru5P and Xu5P are converted into glyceraldehyde-3-phosphate (G3P/GAP), and fructose-6-phosphate (F6P) via the pentose phosphate pathway (PPP) [12] . The PPP is a fundamental component of cellular metabolism that has a purpose of reducing molecules for anabolism and reducing oxidative stress [13] . The enzymes used in the PPP are transketolase and transaldolase. Transketolase and the same starting intermediate, Ru5P, can be used start the Sedoheptulose-1,7-biphosphate pathways which, also using PPi-PFK and FBP adolase, produce the same products as PPP. These products, F6P and G3P can enter the glycolysis cycle to be further oxidized using the DHAP anion. The product of glycolysis is pyruvate. Figure 4 shows a diagram of these pathways [12].
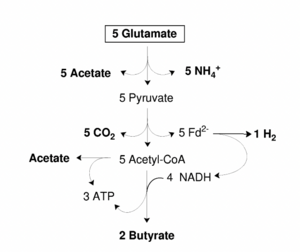
Byruvate formation takes place in the large intestine mostly vis gram-positive anaerobic firmicutes. The two most abundant groups in the large intestine are Eubacterium rectale/ Roseburia spp. and to Faecalibacterium prausnitzii. Byruvate is an important component of the large intestine because it is a main energy source for colonic mucosa and regulates gene expression, inflammation, and apoptosis. More specifically, it can activate the AP-1 signaling pathway in epithial cells by interacting dirst with human cell receptors FFAR2 and FFAR3. The AP-1 pathway controls a number of cellular processes such as cellular differentiation, proliferation, and apoptosis. This interaction can protect against diet-induced obesity by improving insulin sensitivity. Butyrate-producing bacteria carry out fermentative metabolism and can also produce lactate, formate, hydrogen and carbon dioxide. A hypothetical scheme of butyrate production is shown in figure 5. It was proposed by Herman et al in 2008 that as well as regenerating NAD+ from NADH, that the production of butyrate may also produce a proton motive force with NADH and ferredoxin oxidoreductase [14] . An example of a specific bacteria that plays a major role in the degradation of xylan is Roseburia intestinalis. This bacterium, as well as E. rectale thrive at a pH of 5.5. The large intestine microbiome is heavily dependent on the hosts diet since much of the microbe’s energy source is dependent on chyme. Resistant starch intake increases the butyrate production in the bacteria [15].
Vitamin B Production in the Large Intestine
The production of vitamin B is also credited to colonic microbes. The human body only needs a very small dose (about 1 ug/day) to help with two enzymes, methionine synthase and (R)-methylmalonyl-CoA mutase [16]. In the small intestine alkaline phosphatase converts thiamine pyrophosphate (TPP), which is the form of vitamin B1 found in lots of meats, eggs, rice bran, and beans, into free thiamine. [16] Thiamine transporters then assist to the intestinal epithelium to absorb the free thiamine which then enters the bloodstream and gets delivered to the necessary places throughout the body. This thiamine is reverted back to TPP to be used for the TCA cycle as energy [17].
Food is a major source of vitamin B1 intake. However, the microbiota of the human colon is also capable of producing it. The human colon holds a diverse microbiota that produces vitamin B1, B2, B3, B4, B5, B6, B7, B9, and B12. The vitamin B1 producers are Bacteroides fragilis and Prevotella copri (phylum Bacteroidetes); Clostridium difficile, some Lactobacillus spp., and Ruminococcus lactaris (Firmicutes); Bifidobacterium spp. (Actinobacteria); and Fusobacterium varium [17]. These bacteria contain genomes that encode for enzymes that synthesize precursors to thiamine, particularly TPP [18]. These precursors are also responsible for the production of thiazole and pyrimidine. Because of the lack of alkaline phosphatase in the colon, free thiamine cannot be produced. So, TPP transporters help the colon absorb the TPP which then enters mitochondria which is used for ATP generation. This energy goes to the colon. However, there is some competition for vitamin B1 in the colon because some firmicutes, such as Faecalibacterium spp., lack the full synthesis pathway for vitamin B1. Faecalibacterium live in the colon and do not produce vitamin B1, but actually require it for growth. Therefore, they must compete with the human colon for it [17].
Vitamin B12, like vitamin B1, can be either produced in the colon or absorbed from food in the small intestine. Vitamin B12 is an important factor, along with vitamin B6 and B9 to form red blood cells and nucleic acids. Vitamin B12 from food sources is absorbed into the epithelial cells of the small intestine via an intrinsic factor. It is produced in the colon from Bacteroides fragilis and Prevotella copri (Bacteroidetes); Clostridium difficile, Faecalibacterium prausnitzii and Ruminococcus lactaris (Firmicutes); Bifidobacterium animalis, B.infantis, and B.longum (Actinobacteria); Fusobacterium varium (Fusobacteria). These bacteria are capable of this production via either an aerobic or anaerobic process [19].
The gut microbiota contains significantly more enzymes that are able to digest food than the human genome contains. So, having gut bacteria is critical to a healthy human. The beginning parts of the GI tract help to break down food and digest what the human enzymes are capable to metabolizing, and then once the chyme enters the large intestine the majority of the microbes take over. The body only excretes what both the human and microbial enzymes are unable to digest. A healthy diet helps to sustain a diverse microbial environment; however, some bacteria can over grow in the gut and cause problems to the host. H. pylori is an example of one bacterium that can overgrow in the stomach and cause gastritis. However, there are other bacteria as well that can even be fatal such as Clostridium difficile (C. diff). C. difficile can be commonly found in healthy infants but should be undetectable in healthy adults [20]. This is one example of how bacteria can be beneficial in some environments and harmful in others. Depending on the environment some bacteria that may be beneficial is very small amount may be very harmful in large amounts. A diverse gut microbiota helps to keep each species in check to meet the needs of both the host and themselves.
References
- ↑ 1.0 1.1 Morowitz MJ, Carlisle EM, Alverdy JC. Contributions of intestinal bacteria to nutrition and metabolism in the critically ill. Surg Clin North Am. 2011 Aug;91(4):771-85, viii. doi: 10.1016/j.suc.2011.05.001. PMID: 21787967; PMCID: PMC3144392.
- ↑ “Illustration of the Digestive System Labeled.” National Institute of Diabetes and Digestive and Kidney Diseases, U.S. Department of Health and Human Services, https://www.niddk.nih.gov/news/media-library/18162.
- ↑ “Your Digestive System & How It Works - Niddk.” National Institute of Diabetes and Digestive and Kidney Diseases, U.S. Department of Health and Human Services, https://www.niddk.nih.gov/health-information/digestive-diseases/digestive-system-how-it-works.
- ↑ 4.0 4.1 Nardone, Gerardo, and Debora Compare. “The Human Gastric Microbiota: Is It Time to Rethink the Pathogenesis of Stomach Diseases?” United European Gastroenterology Journal, vol. 3, no. 3, 2015, pp. 255–260., https://doi.org/10.1177/2050640614566846.
- ↑ 5.0 5.1 5.2 Algood HM, Cover TL. Helicobacter pylori persistence: an overview of interactions between H. pylori and host immune defenses. Clin Microbiol Rev. 2006 Oct;19(4):597-613. doi: 10.1128/CMR.00006-06. PMID: 17041136; PMCID: PMC1592695.
- ↑ Bik, Elisabeth M., et al. “Molecular Analysis of the Bacterial Microbiota in the Human Stomach.” Proceedings of the National Academy of Sciences, vol. 103, no. 3, 2006, pp. 732–737., https://doi.org/10.1073/pnas.0506655103.
- ↑ 7.0 7.1 Sahar El Aidy, Bartholomeus van den Bogert, Michiel Kleerebezem. The small intestine microbiota, nutritional modulation and relevance for health. Current Opinion in Biotechnology. Volume 32. 2015. Pages 14-20. ISSN 0958-1669. https://doi.org/10.1016/j.copbio.2014.09.005.
- ↑ Van den Bogert, Bartholomeus, et al. “Diversity of Human Small Intestinal Streptococcu sand Veillonella Populations.” FEMS Microbiology Ecology, vol. 85, no. 2, 2013, pp. 376–388., https://doi.org/10.1111/1574-6941.12127.
- ↑ Zoetendal, Erwin G, et al. “The Human Small Intestinal Microbiota Is Driven by Rapid Uptake and Conversion of Simple Carbohydrates.” The ISME Journal, vol. 6, no. 7, 2012, pp. 1415–1426., https://doi.org/10.1038/ismej.2011.212.
- ↑ Hooper LV, Xu J, Falk PG, Midtvedt T, Gordon JI. A molecular sensor that allows a gut commensal to control its nutrient foundation in a competitive ecosystem. Proc Natl Acad Sci U S A. 1999 Aug 17;96(17):9833-8. doi: 10.1073/pnas.96.17.9833. PMID: 10449780; PMCID: PMC22296.
- ↑ Azzouz LL, Sharma S. Physiology, Large Intestine. [Updated 2022 Aug 1. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2023 Jan-. Available from: https://www.ncbi.nlm.nih.gov/books/NBK507857/.]
- ↑ 12.0 12.1 12.2 Garschagen, Laura S., et al. “An Alternative Pentose Phosphate Pathway in Human Gut Bacteria for the Degradation of C5 Sugars in Dietary Fibers.” The FEBS Journal, vol. 288, no. 6, 2020, pp. 1839–1858., https://doi.org/10.1111/febs.15511.
- ↑ Bartlett et al.: Oncolytic viruses as therapeutic cancer vaccines. Molecular Cancer 2013 12:103.
- ↑ Herrmann G, Jayamani E, Mai G, Buckel W. Energy conservation via electron-transferring flavoprotein in anaerobic bacteria. J Bacteriol. 2008 Feb;190(3):784-91. doi: 10.1128/JB.01422-07. Epub 2007 Nov 26. PMID: 18039764; PMCID: PMC2223574.
- ↑ Russell, Wendy R, et al. “Colonic Bacterial Metabolites and Human Health.” Current Opinion in Microbiology, vol. 16, no. 3, 2013, pp. 246–254., https://doi.org/10.1016/j.mib.2013.07.002.
- ↑ 16.0 16.1 Martens, H. Barg, M. Warren, D. Jah, J.-H. “Microbial Production of Vitamin B 12.” Applied Microbiology and Biotechnology, vol. 58, no. 3, 2002, pp. 275–285., https://doi.org/10.1007/s00253-001-0902-7.
- ↑ 17.0 17.1 17.2 Yoshii K, Hosomi K, Sawane K, Kunisawa J. Metabolism of Dietary and Microbial Vitamin B Family in the Regulation of Host Immunity. Front Nutr. 2019 Apr 17;6:48. doi: 10.3389/fnut.2019.00048. PMID: 31058161; PMCID: PMC6478888.
- ↑ Putnam, Emily E., and Andrew L. Goodman. “B Vitamin Acquisition by Gut Commensal Bacteria.” PLOS Pathogens, vol. 16, no. 1, 2020, https://doi.org/10.1371/journal.ppat.1008208.
- ↑ Fang, Huan, et al. “Microbial Production of Vitamin B12: A Review and Future Perspectives.” Microbial Cell Factories, vol. 16, no. 1, 2017, https://doi.org/10.1186/s12934-017-0631-y.
- ↑ “Peptoclostridium Difficile.” Peptoclostridium Difficile - an Overview | ScienceDirect Topics, https://www.sciencedirect.com/topics/medicine-and-dentistry/peptoclostridium-difficile.
Authored for BIOL 238 Microbiology, taught by Joan Slonczewski, 2023, Kenyon College
