Panteoa agglomerans: Difference between revisions
| Line 51: | Line 51: | ||
==References== | ==References== | ||
Al-Kidsawey, E. R., Mhdi, K. H., & Al kaif, M. A. (2020). Isolation and identification of Pantoea agglomerans from open heart operations unit of Marjan Hospital in Hilla City. Indian Journal of Forensic Medicine & Toxicology. https://doi.org/10.37506/ijfmt.v14i1.129 | |||
Al-Qaysi, S. A., Al-Haideri, H., Al-Shimmary, S. M., Abdulhameed, J. M., Alajrawy, O. I., Al-Halbosiy, M. M., Moussa, T. A., & Farahat, M. G. (2021). Bioactive Levan-Type Exopolysaccharide produced by pantoea agglomerans ZMR7: Characterization and optimization for enhanced production. Journal of Microbiology and Biotechnology, 31(5), 696–704. https://doi.org/10.4014/jmb.2101.01025 | |||
Dillon, R. J., Vennard, C. T., & Charnley, A. K. (2000). Exploitation of gut bacteria in the Locust. Nature, 403(6772), 851–851. https://doi.org/10.1038/35002669 | |||
Dutkiewicz, J., Mackiewicz, B., Lemieszek, M. K., Golec, M., & Milanowski, J. (2016). pantoea agglomerans: A mysterious bacterium of evil and good. part IV. beneficial effects. Annals of Agricultural and Environmental Medicine, 23(2), 206–222. https://doi.org/10.5604/12321966.1203879 | |||
Dutkiewicz, J., Mackiewicz, B., Lemieszek, M., Golec, M., & Milanowski, J. (2015). pantoea agglomerans: A marvelous bacterium of evil and good. part I. deleterious effects: dust-borne endotoxins and allergens – focus on cotton dust. Annals of Agricultural and Environmental Medicine, 22(4), 576–588. https://doi.org/10.5604/12321966.1185757 | |||
Guevarra, R. B., Magez, S., Peeters, E., Chung, M. S., Kim, K. H., & Radwanska, M. (2020). Comprehensive Genomic Analysis Reveals Virulence Factors and Antibiotic Resistance Genes in Pantoea Agglomerans KM1, a Potential Opportunistic Pathogen. https://doi.org/10.1101/2020.09.15.297663 | |||
Kaur, I. P., Inkollu, S., Prakash, A., Gandhi, H., Mughal, M. S., & Du, D. (2020). pantoea agglomerans bacteremia: Is it dangerous? Case Reports in Infectious Diseases, 2020, 1–4. https://doi.org/10.1155/2020/7890305 | |||
Lee, S. I., Tran, T. D., Huynh, S., Parker, C. T., Hnasko, R., & McGarvey, J. A. (2021). Complete genome sequence of Pantoea agglomerans ASB05 using Illumina and PacBio sequencing. Microbiology Resource Announcements, 10(30). https://doi.org/10.1128/mra.00501-21 | |||
Li, L., Chen, R., Zuo, Z., Lv, Z., Yang, Z., Mao, W., Liu, Y., Zhou, Y., Huang, J., & Song, Z. (2020). Evaluation and improvement of phosphate solubilization by an isolated bacterium pantoea agglomerans zb. World Journal of Microbiology and Biotechnology, 36(2). https://doi.org/10.1007/s11274-019-2744-4 | |||
Lorenzi, A. S., Bonatelli, M. L., Chia, M. A., Peressim, L., & Quecine, M. C. (2022). Opposite sides of pantoea agglomerans and its associated commercial outlook. Microorganisms, 10(10), 2072. https://doi.org/10.3390/microorganisms10102072 | |||
Matsuzawa, T., Mori, K., Kadowaki, T., Shimada, M., Tashiro, K., Kuhara, S., Inagawa, H., Soma, G. -i., & Takegawa, K. (2012). Genome sequence of Pantoea agglomerans strain IG1. Journal of Bacteriology, 194(5), 1258–1259. https://doi.org/10.1128/jb.06674-11 | |||
Morin, A. (2014). Pantoea. Encyclopedia of Food Microbiology, 1028–1032. https://doi.org/10.1016/b978-0-12-384730-0.00245-7 | |||
Morin, André, & Parveen, Z. (1999). Pantoea. Encyclopedia of Food Microbiology, 1623–1630. https://doi.org/10.1006/rwfm.1999.1220 | |||
Pesticide Programs, Pantoea agglomerans strain C9-1 1–30 (2009). Ottawa; Pest Management Regulatory Agency. | |||
U.S. National Library of Medicine. (n.d.). Taxonomy browser (pantoea agglomerans IG1). National Center for Biotechnology Information. | |||
https://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Info&id=1110694&lvl=3&lin=f&keep=1&srchmode=1&unlock | |||
Walterson, A. M., & Stavrinides, J. (2015). pantoea:insights into a highly versatile and diverse genus within the Enterobacteriaceae. FEMS Microbiology Reviews, 39(6), 968–984. https://doi.org/10.1093/femsre/fuv027 | |||
Wang, S., Ghosh, A. K., Bongio, N., Stebbings, K. A., Lampe, D. J., & Jacobs-Lorena, M. (2012). Fighting malaria with engineered symbiotic bacteria from Vector Mosquitoes. Proceedings of the National Academy of Sciences, 109(31), 12734–12739. https://doi.org/10.1073/pnas.1204158109 | |||
==Author== | ==Author== | ||
Revision as of 02:14, 12 December 2023
Classification
Bacteria, Pseudomonadota, Gammaproteobacteria, Enterobacterales, Erwiniaceae, Pantoea
Species
|
NCBI: [1] |
Pantoea agglomerans
Description and Significance
Pantoea agglomerans was first described in 1888 and has been identified under many names including: Enterobacter agglomerans, Erwinia herbicola, Bacillus agglomerans, Bacterium herbicola, Pseudomonas herbicola, Agrobacterium gypsophilae, Bacterium typhi flavum, Erwinia lathyr, Corynebacterium beticola, Flavobacterium rhenanum, Pseudomonas trifolii, and Xanthomonas trifolii.
The etymology of its name is ‘Pantoea’, derived from the Greek word ‘pantoios’, which means ‘of all sorts and sources’, reflecting its ubiquitous occurrence; and ‘agglomerans’ meaning ‘forming into a ball’, due to the tendency for P. agglomerans to aggregate into clusters.
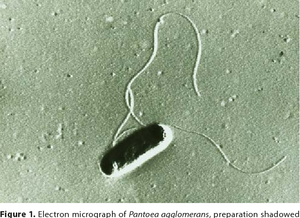
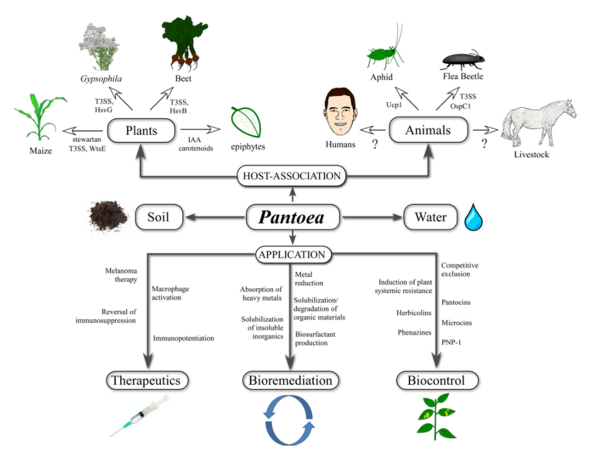
Pantoea agglomerans is a non-spore forming, Gram-negative, rod-shaped, facultative anaerobe bacillus that is motile via peritrichous flagella. It forms yellow pigmented colonies with 5mm concave centers. P. agglomerans grows quickly on agar at 30°C at a rate comparable to E. coli. P. agglomerans is found ubiquitously on plants as both an epiphyte and an endophyte. It forms host associations with a diverse variety of hosts including plants, insects, and humans. P. agglomerans is found abundantly in plants, animals, arthropods, feces, soil, water, dust, air, and in humans. P. agglomerans is as ubiquitous in function and application as it is in occurrence, it produces a wide variety of molecules depending on where it is grown with both positive and negative attributes and wide applications for bioremediation, biocontrol, and therapeutics.
P. agglomerans has multiple bioremediation applications as a natural fertilizer and pesticide, replacing harmful chemical alternatives. Other bioremediation applications include natural feed additives for poultry, breaking down herbicides into nontoxic byproducts, and the ability to break down petroleum hydrocarbons. P. agglomerans demonstrates a wide array of medical applications as well, producing multiple compounds that are utilized to produce powerful biocontrol agents for humans and plants that function as antibiotics, antifungals, and antivirals. P. agglomerans also produces a unique lipopolysaccharide that acts as a macrophage activator and immunopotentiator which has been utilized in research to suppress cancerous tumors. Overall, P. agglomerans is an incredibly useful bacteria that is easy and quick to grow with a wide variety of integral apllications.
Genome Structure
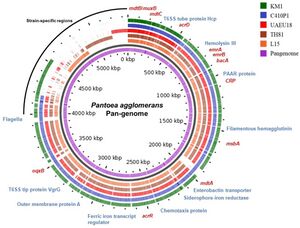
P. agglomerans has a single circular genome 4.8Mb long with a median protein count of 4,435 and a GC content of 55%. The chromosome contains 3-5 large plasmids ranging from 280-789 kb as well as 3,615 coding sequences, 77 tRNA genes and 22 rRNA operons. The genome has been fully sequenced to 76-fold coverage by a whole-genome shotgun strategy. One shotgun and 0.5 paired-end reads were performed using a Roche 454 GS (FLX titanium) pyrosequencer. All of the reads were assembled using Newbler Assembler 2.5 (454 Life Science), which generated 18 large contigs (>500 bp).
P. agglomerans has been identified as a cause of human infection by utilizing PCR techniques to identify the repA gene, which is linked to plant pathogenicity. P. agglomerans is symbiotic with locust species, producing hpc and vgrG, which code for effector proteins that make vital hormones. It also contains quorum sensing genes such as acyl-homoserine lactones.
Cell Structure, Metabolism and Life Cycle
Pantoea agglomerans is a facultative anaerobe that utilizes atmospheric nitrogen and converts it into ammonia. P. agglomerans aggregates into elongated, sausage shaped colonies that form a translucent exopolysaccharide symplasmata.
P. agglomerans produces a wide variety of molecules that have been utilized in many ways to create effective fertilizers, medicines, and biocontrol agents among other compounds. It is also a key microbe to numerous plant and insect symbionts.
P. agglomerans’’ produces multiple organic acids that solubilizes phosphorus and potassium as well as folic acid which is a crucial step in the fermentation process of rye bread, prior to the growth of the lactobacilli. Another acid produced by P. agglomerans is α-hydroxy-4-methylthiobutyric acid, which is utilized as a natural feed additive for poultry due to its amino acids that contain a high sulfur content. P. agglomerans also produces several hormones that are crucial to plant and insect symbionts. It is a key part of the gut biota in many insects including locusts, which depend on P. agglomerans to produce phenol and guaiacol. These are utilized to produce a hormone that signals locusts to aggregate together in swarms. P. agglomerans also produces several phytohormones that wheat species depend on for nitrogen fixation and nitrate reduction.
P. agglomerans produces various molecules with versatile anthropogenic uses. P. agglomerans produces a lipase that is effective in reducing lipids. It interchanges the hypercholesterolaemic fatty acids into hyporcholesterolaemic fatty acids in butter, creating a more healthy alternative. P. agglomerans is also capable of converting nitrile into industrial amides such as nicotinamide, which is utilized in the creation of vitamins. Another compound synthesized from P. agglomerans is a glycolipid biosurfactant capable of degrading petroleum hydrocarbons that is particularly effective in contaminated soil.
P. agglomerans is valued for its ability to create antagonistic compounds that can be used to form biocontrol agents, activate enzymes, and detoxify substances. It is used in the synthesis of many antibiotics, antifungals, antiparasitics, and antivirals including Heribcolin, Phenazine, pantocins, microcin, agglomerans and andrimid. There are many promising medical applications to compounds synthesized by P. agglomerans. When grown in maple syrup, P. agglomerans produces a levan, an exopolysaccharide polymer. This polymer acts as a food stabilizer that could replace xanthan as a healthier alternative. The levan produced by P. agglomerans also has promising medical applications and has been shown to inhibit the growth of cancers such as sarcoma and breast cancer. P. agglomerans has a unique low molecular mass lipopolysaccharide, IP-PA1, that is being researched for its ability to stimulate macrophage activity in targeted areas. A promising study from the Institute of Rural Health in Lublin, Poland revealed that the immunopotentiating qualities of the P. agglomerans lipopolysaccharide cause a total suppression of malignant tumors when administered transdermally or orally to mice. In addition to acting as effective tumor suppressors, the lipopolysaccharides have anticholesterolaemic, antiulcer, antihemorrhoidal, antidiabetic, antihyperlipidemia, antiallergy, phylactic, and analgesic effects. Furthermore the lipopolysaccharide promotes egg laying in poultry and aids in the upregulation of bone turnover.
Ecology and Pathogenesis
Habitat; symbiosis; biogeochemical significance; contributions to environment.
If relevant, how does this organism cause disease? Human, animal, plant hosts? Virulence factors, as well as patient symptoms.
References
Al-Kidsawey, E. R., Mhdi, K. H., & Al kaif, M. A. (2020). Isolation and identification of Pantoea agglomerans from open heart operations unit of Marjan Hospital in Hilla City. Indian Journal of Forensic Medicine & Toxicology. https://doi.org/10.37506/ijfmt.v14i1.129
Al-Qaysi, S. A., Al-Haideri, H., Al-Shimmary, S. M., Abdulhameed, J. M., Alajrawy, O. I., Al-Halbosiy, M. M., Moussa, T. A., & Farahat, M. G. (2021). Bioactive Levan-Type Exopolysaccharide produced by pantoea agglomerans ZMR7: Characterization and optimization for enhanced production. Journal of Microbiology and Biotechnology, 31(5), 696–704. https://doi.org/10.4014/jmb.2101.01025
Dillon, R. J., Vennard, C. T., & Charnley, A. K. (2000). Exploitation of gut bacteria in the Locust. Nature, 403(6772), 851–851. https://doi.org/10.1038/35002669
Dutkiewicz, J., Mackiewicz, B., Lemieszek, M. K., Golec, M., & Milanowski, J. (2016). pantoea agglomerans: A mysterious bacterium of evil and good. part IV. beneficial effects. Annals of Agricultural and Environmental Medicine, 23(2), 206–222. https://doi.org/10.5604/12321966.1203879
Dutkiewicz, J., Mackiewicz, B., Lemieszek, M., Golec, M., & Milanowski, J. (2015). pantoea agglomerans: A marvelous bacterium of evil and good. part I. deleterious effects: dust-borne endotoxins and allergens – focus on cotton dust. Annals of Agricultural and Environmental Medicine, 22(4), 576–588. https://doi.org/10.5604/12321966.1185757
Guevarra, R. B., Magez, S., Peeters, E., Chung, M. S., Kim, K. H., & Radwanska, M. (2020). Comprehensive Genomic Analysis Reveals Virulence Factors and Antibiotic Resistance Genes in Pantoea Agglomerans KM1, a Potential Opportunistic Pathogen. https://doi.org/10.1101/2020.09.15.297663
Kaur, I. P., Inkollu, S., Prakash, A., Gandhi, H., Mughal, M. S., & Du, D. (2020). pantoea agglomerans bacteremia: Is it dangerous? Case Reports in Infectious Diseases, 2020, 1–4. https://doi.org/10.1155/2020/7890305
Lee, S. I., Tran, T. D., Huynh, S., Parker, C. T., Hnasko, R., & McGarvey, J. A. (2021). Complete genome sequence of Pantoea agglomerans ASB05 using Illumina and PacBio sequencing. Microbiology Resource Announcements, 10(30). https://doi.org/10.1128/mra.00501-21
Li, L., Chen, R., Zuo, Z., Lv, Z., Yang, Z., Mao, W., Liu, Y., Zhou, Y., Huang, J., & Song, Z. (2020). Evaluation and improvement of phosphate solubilization by an isolated bacterium pantoea agglomerans zb. World Journal of Microbiology and Biotechnology, 36(2). https://doi.org/10.1007/s11274-019-2744-4
Lorenzi, A. S., Bonatelli, M. L., Chia, M. A., Peressim, L., & Quecine, M. C. (2022). Opposite sides of pantoea agglomerans and its associated commercial outlook. Microorganisms, 10(10), 2072. https://doi.org/10.3390/microorganisms10102072
Matsuzawa, T., Mori, K., Kadowaki, T., Shimada, M., Tashiro, K., Kuhara, S., Inagawa, H., Soma, G. -i., & Takegawa, K. (2012). Genome sequence of Pantoea agglomerans strain IG1. Journal of Bacteriology, 194(5), 1258–1259. https://doi.org/10.1128/jb.06674-11
Morin, A. (2014). Pantoea. Encyclopedia of Food Microbiology, 1028–1032. https://doi.org/10.1016/b978-0-12-384730-0.00245-7
Morin, André, & Parveen, Z. (1999). Pantoea. Encyclopedia of Food Microbiology, 1623–1630. https://doi.org/10.1006/rwfm.1999.1220 Pesticide Programs, Pantoea agglomerans strain C9-1 1–30 (2009). Ottawa; Pest Management Regulatory Agency.
U.S. National Library of Medicine. (n.d.). Taxonomy browser (pantoea agglomerans IG1). National Center for Biotechnology Information. https://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Info&id=1110694&lvl=3&lin=f&keep=1&srchmode=1&unlock
Walterson, A. M., & Stavrinides, J. (2015). pantoea:insights into a highly versatile and diverse genus within the Enterobacteriaceae. FEMS Microbiology Reviews, 39(6), 968–984. https://doi.org/10.1093/femsre/fuv027
Wang, S., Ghosh, A. K., Bongio, N., Stebbings, K. A., Lampe, D. J., & Jacobs-Lorena, M. (2012). Fighting malaria with engineered symbiotic bacteria from Vector Mosquitoes. Proceedings of the National Academy of Sciences, 109(31), 12734–12739. https://doi.org/10.1073/pnas.1204158109
Author
Page authored by Samuel Rogers, student of Prof. Bradley Tolar at UNC Wilmington. [Category:Pages edited by students of Bradley Tolar at UNC Wilmington]]