Metallosphaera yellowstonensis: Difference between revisions
| Line 33: | Line 33: | ||
''M. yellowstonensis'' possess the largest genome of the known ''Mettallosphaera'' speceis at 2.82 Mb. Throughout the genus, GC contents range from 42.0-50.4%. (wang et.al) | ''M. yellowstonensis'' possess the largest genome of the known ''Mettallosphaera'' speceis at 2.82 Mb. Throughout the genus, GC contents range from 42.0-50.4%. (wang et.al) | ||
There are ~283 transposon sequences per genome found in ''M. yellowstonensis''. These sequences can provide extra functionalities though HGT events and extra protection | There are ~283 transposon sequences per genome found in ''M. yellowstonensis''. These sequences can provide extra functionalities though HGT events and extra protection from the danger of the hot spring environment. (wang et. al) | ||
==Cell Structure, Metabolism and Life Cycle== | ==Cell Structure, Metabolism and Life Cycle== | ||
Revision as of 13:21, 23 April 2024
Classification
Archaea (Domain); (Superphylum) TACK group "Crenarchaeota"; Thermoproteota (Phylum); Thermoprotei (Class); Sulfolobales (Order); Sulfolobaceae (Family); Metallosphaera (Genus)
 *idk why this one is being weird, also feel free to move the other ones/format it better i thought id just throw them in for now*
*idk why this one is being weird, also feel free to move the other ones/format it better i thought id just throw them in for now*
Species
|
NCBI: [1] |
Description and Significance
This is a coccus-shaped chemolithoautotrophic archaea isolated from the hot springs of Yellowstone National Park (Kozubal, et. al). M. yellowstonensis exists in Fe(II)-oxidizing microbial mats. This microorganism has implications for the origin of eukaryotes as well as insight into unique metabolic pathways in extreme environments (Jennings, et. al).
Thermophilic, acidophilic chemolithoautotrophic bacteria such as M. yellowstonensis are important microorganisms in understanding evolutionary history of the Earth. The ability to survive in Fe (II)-oxidizing mats with minimal nutrient requirements other than inorganic compounds suggests an important role as a primary producer in the extreme environment in which it is found. (Kozubal et.al 2008).
Genome Structure
Describe the size and content of the genome. How many chromosomes? Circular or linear? Other interesting features? What is known about its sequence?
M. yellowstonensis is an archaea, thus its genome resembles that of bacteria in that is circular. Most genes throughout the genome average around 1kbp in length. Additionally, these genes tend to be adjacent to neighboring genes/separated by less than 200bp. This results in high density coding regions and minimal noncoding regions. ........ https://www.sciencedirect.com/topics/biochemistry-genetics-and-molecular-biology/archaeal-genome
M. yellowstonensis possess the largest genome of the known Mettallosphaera speceis at 2.82 Mb. Throughout the genus, GC contents range from 42.0-50.4%. (wang et.al)
There are ~283 transposon sequences per genome found in M. yellowstonensis. These sequences can provide extra functionalities though HGT events and extra protection from the danger of the hot spring environment. (wang et. al)
Cell Structure, Metabolism and Life Cycle
Interesting features of cell structure; how it gains energy; what important molecules it produces.
M. yellowstonesis can utilize different sulfur compounds (sulfide, elemental sulfur, thiosulfate) derived from hotsprings and continental solfataras as an energy source. Additionally, M. yellowstonesis is capable of iron oxidation (fox genes), and also posses an abundant amount of carbohydrate active enzymes that encode for: glycolysis, gluconeogenesis, archaeal pentose phosphate pathway, an atypical TCA cycle, and complete non-phosphative and semi phosphorylative entner doudoroff pathways (Wang et.al).
M. yellowstonensis is unable to fix CO2 or CO to obtain carbon, and thus this must be obtained from autotrophic organisms present in the spring (Kozubal et.al).
Additionally, M. yellowstonesis has putative type I carbon monoxide dehydrogenase. M. yellowstonesis can also perform assimilatory nitrate reduction, with genes for nitrate and nitrite reductases. Unique from the rest of the genus, M. yellowstonensis MK1 also possesses an operon encoding for dissimilatory nitrate reductase. (wang)
Ecology and Pathogenesis
Habitat; symbiosis; biogeochemical significance; contributions to environment.
If relevant, how does this organism cause disease? Human, animal, plant hosts? Virulence factors, as well as patient symptoms.
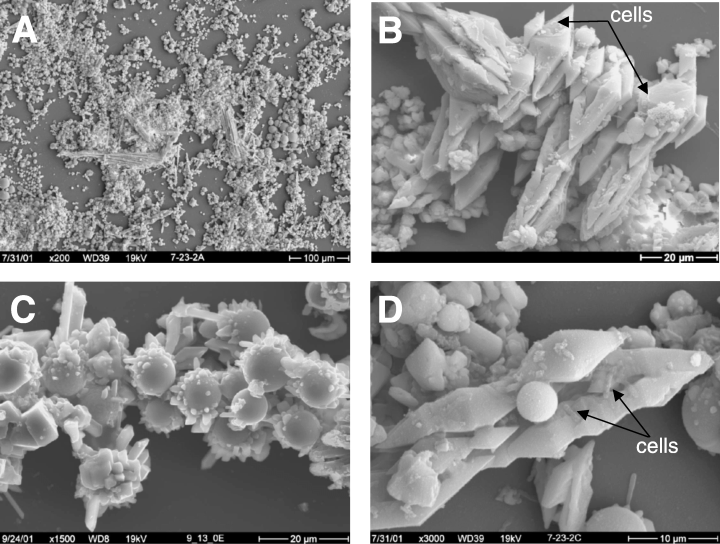
M. yellowstonensis is found in microbial mats (acidic ferric iron mats), which are highly diverse communities that can provide an extreme environment with low pH, high temperatures, low amounts of oxygen, and high concentrations of reduced iron (Kozubal et.al) Different organisms create the ribbons of color seen in the mats. In these mats millions of microbes can connect into long fillaments, or thick sturdy structures coated by chemical precipitates. https://www.nps.gov/yell/learn/nature/thermophilic-communities.htm
M. yellowstonensis produces EPS, which can be utilized in biofilm formation, and adhesion generally assisting in colonization, solubizing minerals, and increased protection form the environment. It also maintains a unique flagellum composition/mode of assembly different from that of bacteria found in the crenarchael flagellin and accessory proteins.
M. yellowstonensis has the ability to survive in natural/anthropogenic metal-rich environments. Unique from its genus, yellowstonensis posesses an alkylmercury lyase, which is important for mercury detoxification.
References
The archaeal ‘TACK’ superphylum and the origin of eukaryotes, https://www.sciencedirect.com/science/article/pii/S0966842X11001740?via%3Dihub
Guy, L., & Thijs J.G. Ettema. (2011). The archaeal “TACK” superphylum and the origin of eukaryotes. Trends in Microbiology (Regular Ed.), 19(12), 580–587. https://doi.org/10.1016/j.tim.2011.09.002
taxonomy. (2020). Taxonomy browser (Metallosphaera yellowstonensis). Nih.gov. https://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Undef&id=1111107&lvl=3&lin=s&keep=1&srchmode=1&unlock&log_op=lineage_toggle
Linking geochemical processes with microbial community analysis: successional dynamics in an arsenic-rich, acid-sulphate-chloride geothermal spring, https://onlinelibrary.wiley.com/doi/full/10.1111/j.1472-4677.2004.00032.x
Macur, R. E., Langner, H. W., Kocar, B. D., & Inskeep, W. P. (2004). Linking geochemical processes with microbial community analysis: successional dynamics in an arsenic‐rich, acid‐sulphate‐chloride geothermal spring. Geobiology, 2(3), 163–177. https://doi.org/10.1111/j.1472-4677.2004.00032.x
Summary of Metallosphaera yellowstonensis MK1, version 28.0. https://biocyc.org/GCF_000243315/organism-summary
Summary of Metallosphaera yellowstonensis MK1, version 28.0. (2022). Biocyc.org. https://biocyc.org/GCF_000243315/organism-summary
General archaeal info: https://www.sciencedirect.com/topics/biochemistry-genetics-and-molecular-biology/archaeal-genome
Agustín Vioque, & Altman, S. (2001). Ribonuclease P. Elsevier EBooks, 137–154. https://doi.org/10.1016/b978-008043408-7/50030-7
Wang article: https://www.frontiersin.org/journals/microbiology/articles/10.3389/fmicb.2020.01192/full Kozubal, M., Macur, R. E., Korf, S., Taylor, W. P., Ackerman, G. G., Nagy, A., & Inskeep, W. P. (2008). Isolation and Distribution of a Novel Iron-Oxidizing Crenarchaeon from Acidic Geothermal Springs in Yellowstone National Park. Applied and Environmental Microbiology, 74(4), 942–949. https://doi.org/10.1128/aem.01200-07
sequence and shape info: https://www.ncbi.nlm.nih.gov/nuccore/NZ_JH597761
Metallosphaera yellowstonensis MK1 MetMK1scaffold_10, whole genome sho - Nucleotide - NCBI. (2024). Nih.gov. https://www.ncbi.nlm.nih.gov/nuccore/NZ_JH597761
microbial mat stuff: https://www.nps.gov/yell/learn/nature/thermophilic-communities.htm
Thermophilic Communities - Yellowstone National Park (U.S. National Park Service). (2020). Nps.gov. https://www.nps.gov/yell/learn/nature/thermophilic-communities.htm
Detailing early experiments with the mats (kozubal 2008) (ps. its from before our guy was discovered but its by the kozubal author an had a good description about the mats) https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2258575/
Kozubal, M., Macur, R. E., Korf, S., Taylor, W. P., Ackerman, G. G., Nagy, A., & Inskeep, W. P. (2008). Isolation and Distribution of a Novel Iron-Oxidizing Crenarchaeon from Acidic Geothermal Springs in Yellowstone National Park. Applied and Environmental Microbiology, 74(4), 942–949. https://doi.org/10.1128/aem.01200-07
Author
Page authored by _____, student of Prof. Jay Lennon at IndianaUniversity.