Bradyrhizobium: Difference between revisions
| Line 30: | Line 30: | ||
==Genome Structure== | ==Genome Structure== | ||
The genome of ''Rhizobium'' ''sp. NGR234'' has a genome structure much like ''[http://biology.kenyon.edu/Microbial_Biorealm/bacteria/proteobacteria/agrobacterium/Agrobacterium.htm Agrobacterium tumefaciens], ''which comes in three parts. However, while ''Agrobacterium'' ''tumefaciens'' has a circular chromosome, a linear chromosome, and a megaplasmid, ''Rhizobium'' ''sp. NGR234'' has a chromosome 3.5 Mb in length, a megaplasmid of more than 2 Mb (pNGR234''b''), and a smaller plasmid 536,165 bp in length (pNGR234''a'') that carries most of the genes used for symbioses with legumes. The average G-C content of the entire genome is about 61.2 mol %. Most of the assumed coding sequences in the ''Rhizobium sp. NGR234'' genome can be "distributed into functional classes similar to those in ''Bacillus subtilis'', [however,] functions related to transposable elements are more abundant in NGR234" (Viprey'' et al.'' 2000). The genome sequencing of [http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=genomeprj&cmd=Retrieve&dopt=Overview&list_uids=13932 ''Rhizobium etli''] and [http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=genomeprj&cmd=Retrieve&dopt=Overview&list_uids=344 ''Rhizobium leguminosarum bv. viciae 3841''] are currently in progress, but ''R. Leguminosarum bv. viciae ''is know to be 7,751,309 bp and the averge G+C content is 60.86 %. <br /><font size="+1"><font size="3"><br /> The genome of ''Bradyrhizobium'' ''japonicum'' is a single chromosome 9,105,828 bp in length. The average G-C content of the genome is 64.1 mol %. Fifty-two percent of the 8,317 potential protein-coding genes are like genes of known function, 30% of the genes are hypothetical, and 18% have no similarity to any reported genes. In addition, 34% of the genes were like genes in ''Mesorhizobium'' ''loti'' and ''Sinorhizobium'' ''meliloti'', and 23% of the genes were unique to ''Bradyrhizobium'' ''japonicum'' (Kaneko'' et al.''). <br / | The genome of ''Rhizobium'' ''sp. NGR234'' has a genome structure much like ''[http://biology.kenyon.edu/Microbial_Biorealm/bacteria/proteobacteria/agrobacterium/Agrobacterium.htm Agrobacterium tumefaciens], ''which comes in three parts. However, while ''Agrobacterium'' ''tumefaciens'' has a circular chromosome, a linear chromosome, and a megaplasmid, ''Rhizobium'' ''sp. NGR234'' has a chromosome 3.5 Mb in length, a megaplasmid of more than 2 Mb (pNGR234''b''), and a smaller plasmid 536,165 bp in length (pNGR234''a'') that carries most of the genes used for symbioses with legumes. The average G-C content of the entire genome is about 61.2 mol %. Most of the assumed coding sequences in the ''Rhizobium sp. NGR234'' genome can be "distributed into functional classes similar to those in ''Bacillus subtilis'', [however,] functions related to transposable elements are more abundant in NGR234" (Viprey'' et al.'' 2000). The genome sequencing of [http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=genomeprj&cmd=Retrieve&dopt=Overview&list_uids=13932 ''Rhizobium etli''] and [http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=genomeprj&cmd=Retrieve&dopt=Overview&list_uids=344 ''Rhizobium leguminosarum bv. viciae 3841''] are currently in progress, but ''R. Leguminosarum bv. viciae ''is know to be 7,751,309 bp and the averge G+C content is 60.86 %. <br /><font size="+1"><font size="3"><br /> The genome of ''Bradyrhizobium'' ''japonicum'' is a single chromosome 9,105,828 bp in length. The average G-C content of the genome is 64.1 mol %. Fifty-two percent of the 8,317 potential protein-coding genes are like genes of known function, 30% of the genes are hypothetical, and 18% have no similarity to any reported genes. In addition, 34% of the genes were like genes in ''Mesorhizobium'' ''loti'' and ''Sinorhizobium'' ''meliloti'', and 23% of the genes were unique to ''Bradyrhizobium'' ''japonicum'' (Kaneko'' et al.''). <br /><br /> The genome of ''Sinorhizobium'' ''meliloti'' is similar to ''Rhizobium'' <font size="+1"><font size="3">sp. NGR234; it has a 3.65 Mb chromosome, a 1.35 Mb megaplasmid (pSymA), and a 1.68 Mb megaplasmid (pSymB). All three genomic elements contribute in some way to plant symbiosis (Galibert ''et al.'' 2001).<br /> | ||
''Mesorhizobium'' ''loti MAFF303099'' has been sequnced by Rhizobase. The genome of ''M. loti'''s simgle chromosome is 7,036,071 bp long, with 6,752 potential protein-coding genes. The G+C content for ''M. loti'' is 62.7%. M. Loti also has two plasmids that were sequenced. Their names are pMLa and pMLb and their G+C contents are 59.3% and 59.9%, respectively. The genome of pMLa is 351, 911 bp long, and the genome of pMLb is 208,315 bp long. | |||
<br />''Azorhizobium'' has not been genetically sequenced but is known to carry out similar processes to other rhizobia. | |||
==Cell Structure and Metabolism== | ==Cell Structure and Metabolism== | ||
Revision as of 20:51, 1 June 2006
|
NCBI: |
Classification
Higher order taxa:
Bacteria; Proteobacteria; Alphaproteobacteria
Species:
Rhizobium (Family: Rhizobiaceae) Rhizobium arachis; Rhizobium daejeonense; Rhizobium etli; Rhizobium galegae; Rhizobium gallicum; Rhizobium giardinii; Rhizobium hainanense; Rhizobium huautlense; Rhizobium indigoferae; Rhizobium leguminosarum; Rhizobium loessense; Rhizobium lupini; Rhizobium mongolense; Rhizobium phaseoli; Rhizobium sullae; Rhizobium tropici; Rhizobium undicola; Rhizobium yangligense; Rhizobium sp.
Bradyrhizobium (Family: Bradyrhizobiaceae) Bradyrhizobium betae; Bradyrhizobrium canariense; Bradyrhizobium elkanii; Bradyrhizobium genosp.; Bradyrhizobium japonicum; Bradyrhizobium liaonigense; Bradyrhizobium lupini; Bradyrhizobium yuanmingense; Bradyrhizobium sp.
Mesorhizobium (Family: Phyllobacteriaceae): Mesorhizobium amorphae; Mesorhizobium chacoense; Mesorhizobium ciceri; Mesorhizobium huakuii; Mesorhizobium loti; Mesorhizobium mediterraneum; Mesorhizobium plurifarium; Mesorhizobium septentrionale; Mesorhizobium temperatum; Mesorhizobium tianshanense; Mesorhizobium sp.
Sinorhizobium (Family: Rhizobiaceae): Sinorhirobium abri; Sinorhizobium americanus; Sinorhizobium arboris; Sinorhizobium fredii; Sinorhizobium indiaensis; Sinorhizobium kostiense; Sinorhizobium kummerowiae; Sinorhizobium medicae; Sinorhizobium meliloti; Sinorhizobium saheli; Sinorhizobium terangae; Sinorhizobium xinjiangense; Sinorhizobium sp.
Azorhizobium (Family: Hyphomicrobiaceae): Azohirzobium caulinodans; Azohirobium johannae; Azorhizobium sp.
Description and Significance
Rhizobium, Bradyrhizobium, Mesorhizobium, Sinorhizobium, and Azorhizobium - known as rhizobia - are symbiotic nitrogen fixers that can be found in the roots of plants and especially in legume plants. They are responsible for the worlds largest portion of fixed atmospheric nitrogen. (Nitrogen-fixation by organisms provides about 65% of the the biosphere's available nitrogen (Lodwig et al. 2003).) Bradyrhizobium japonicum has been used since 1957 in molecular genetics, physiology, and ecology due to its exellent ability in symbiotic nitrogen fixation.
Genome Structure
The genome of Rhizobium sp. NGR234 has a genome structure much like Agrobacterium tumefaciens, which comes in three parts. However, while Agrobacterium tumefaciens has a circular chromosome, a linear chromosome, and a megaplasmid, Rhizobium sp. NGR234 has a chromosome 3.5 Mb in length, a megaplasmid of more than 2 Mb (pNGR234b), and a smaller plasmid 536,165 bp in length (pNGR234a) that carries most of the genes used for symbioses with legumes. The average G-C content of the entire genome is about 61.2 mol %. Most of the assumed coding sequences in the Rhizobium sp. NGR234 genome can be "distributed into functional classes similar to those in Bacillus subtilis, [however,] functions related to transposable elements are more abundant in NGR234" (Viprey et al. 2000). The genome sequencing of Rhizobium etli and Rhizobium leguminosarum bv. viciae 3841 are currently in progress, but R. Leguminosarum bv. viciae is know to be 7,751,309 bp and the averge G+C content is 60.86 %.
The genome of Bradyrhizobium japonicum is a single chromosome 9,105,828 bp in length. The average G-C content of the genome is 64.1 mol %. Fifty-two percent of the 8,317 potential protein-coding genes are like genes of known function, 30% of the genes are hypothetical, and 18% have no similarity to any reported genes. In addition, 34% of the genes were like genes in Mesorhizobium loti and Sinorhizobium meliloti, and 23% of the genes were unique to Bradyrhizobium japonicum (Kaneko et al.).
The genome of Sinorhizobium meliloti is similar to Rhizobium sp. NGR234; it has a 3.65 Mb chromosome, a 1.35 Mb megaplasmid (pSymA), and a 1.68 Mb megaplasmid (pSymB). All three genomic elements contribute in some way to plant symbiosis (Galibert et al. 2001).
Mesorhizobium loti MAFF303099 has been sequnced by Rhizobase. The genome of M. loti's simgle chromosome is 7,036,071 bp long, with 6,752 potential protein-coding genes. The G+C content for M. loti is 62.7%. M. Loti also has two plasmids that were sequenced. Their names are pMLa and pMLb and their G+C contents are 59.3% and 59.9%, respectively. The genome of pMLa is 351, 911 bp long, and the genome of pMLb is 208,315 bp long.
Azorhizobium has not been genetically sequenced but is known to carry out similar processes to other rhizobia.
Cell Structure and Metabolism
Rhizobium, Bradyrhizobium, Mesorhizobium, Sinorhizobium, and Azorhizobium - collectively known as rhizobia - are Gram-negative, nitrogen-fixing bacteria that form nodules on host plants. They also have symbiotic relationships with legume plants, which can't live without these bacteria's essential nitrogen-fixing processes. In nodules, the rhizobia bacteriods use carbon and energy from the plant in the form of dicarboxylic acids. Recent studies have suggested that the bacteroids do more than just provide the plant with ammonium (through nitrogen fixation). It was shown that a more complex amino-acid cycle is needed for Rhizobium to successfully fix nitrogen in pea nodules. Rhizobium can use the amino acids from the plant to shut down their ammonium assimilation; however, the bacteria must provide the plant with ammonium in order to obtain the amino acids. This alone would mean that the plant could regulate the amount of dicarboxylate that the bacteroids use by amino acid supply and dominate the relationship. This is not the case, however, because the bacteroids "act like plant organelles to cycle amino acids back to the plant for asparagine synthesis," making the plant dependent on them (Lodwig et al. 2003). This system creates mutualism between the bacteria and the plant. | width="399" valign="top" | File:Beans2.jpe
Rhizobium bacteroids in bean nodules. From Philip Poole |}{| width="797" | However, nitrogen fixation is an energy expensive process that requires up to 22% of the plants net photosynthate. In addition, at least 25% of the electron flux through the nitrogenase goes towards reducing protons into hydrogen gas. This process of nitrogenase-dependent hydrogen production is a major factor in the efficiency of symbiotic nitrogen fixation. To have more efficient energy use, some Rhizobium and many Bradyrhizobium strains recycle the hydrogen produced by nitrogenase in nodule bacteroids that have a hydrogen uptake system (Hup). However, Sinorhizobium meliloti, M. ciceri, and R. leguminosarum by. viciae UML2 strains have poor expression of the hup system (Palacios et al. 2000). |}{| width="100%" | width="66%" |
Rhizobium nodules on the roots of a soybean plant. From the US Dept of Agriculture Natural Resources Conservation Service
|}{| width="100%" | width="72%" height="79" valign="top" |
Ecology
Rhizobia can be found in the roots, or rhizosphere, of other types of plants where they cause the formation of nodules. For example, Bradyrhizobium japonicum was first isolated from a soybean nodule in Florida in 1957. Rhizobium sp. NGR234 has a host range of more than 112 genera of legumes (Viprey et al. 2000). These symbiotic relationships occur when rhizobia penetrate their hosts with centripetally-developing infection threads. The bacterium induces the a meristem at the cortex of the plant roots where nodules then develop. Meanwhile, the infection threads make their way into the nodule cells and release rhizobia into the cytoplasm of infected cells. The rhizobia, which act as symbiosomes, enlarge and differentiate into nitrogen-fixing bacteroids, which have low free-oxygen levels. The symbiotic development comes from an exchange of chemical signals between the plant and the bacteria. One of the first signals in this continuous exchange are called flavonoids and are released by the legume roots. They actually activate the expression of nodulation genes (nod, noe, and nol) by interacting with rhizobial regulators of the NodD family. Most of these nodulations genes then help synthesis and secrete a family of lipochito-oligosaccharide molecules that help the bacteria get into the root hairs (Viprey et al. 2000). |}{| width="775" | width="769" |
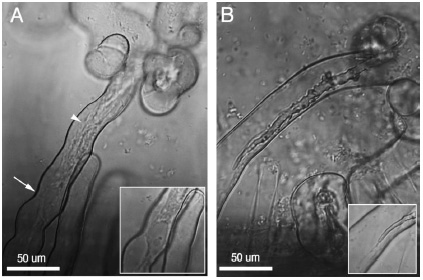
Photograph of infection threads in a root hair. In A, the arrows point to the tip of the infection thread and the nucleus with a column of cytoplasm between them; in B the terminated infection thread as well as the tip of the thread is easily discernable. From Daniel J. Gage
|}
Nodulation
The stages in which nodules form have been named and described in order to better explain the complex system of Rhizobium's interaction with legume plants (visit The Hebrew University of Jerusalem site for in-depth information on the nodulation stages and an article on rhizobia infection and invasion of roots by Daniel J. Gage). {| width="799"
| width="441" height="370" |
| roc | multiplication of rhizobia at or near the root surface |
| roa | adhesion of rhizobia to root hair surface |
| hab and hac | root hair branching and root curling |
| inf | formation of an infection thread |
| noi | nodule initiation: formation of nodule meristem, nodule development and differentiation |
| bar | bacteroid release from infection thread |
| bad | bacterial differentiation |
| nif | onset of nitrogen fixation |
| cof | biochemcial and physiological functions associated with nitrogen fixation |
| nop | maintenance (persistence) of nodule function |
Table showing the stages of nodulation. From The Hebrew University of Jerusalem | width="348" |
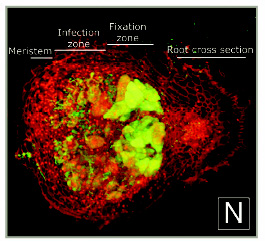
A cross-section of a 10-day-old alfalfa nodule with S. meliloti. Notice the infection thread network behind the meristem region of the nodule. The initial infection site is at the left. Propidium iodide gives the red color to the plant tissue. The root can be seen at the right. From Daniel J. Gage
|}{| width="795"
| width="789" height="328" align="right" valign="top" |
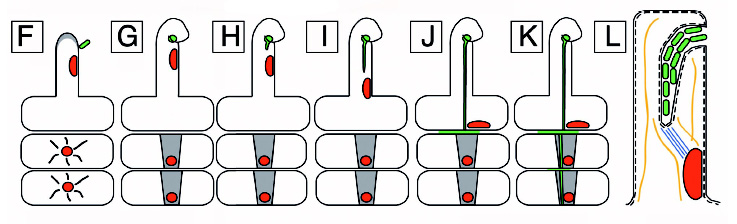
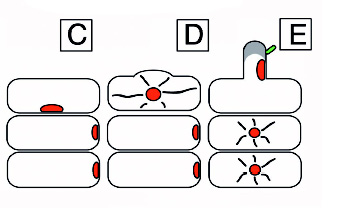
A diagram of nodulation. C) An epidermal cell with a nucleus acroos from where a new root hair will form and two outer cortical cells under it. D) The epidermal cell initiating root hair growth. E) A rhizobial cell binding to a root hair activating the cortical cells in response to the Nod (nodulatin genes) factor. F) Continued growth of the root hair. G) Curling of the root hair due to the Nod factor. The cortical cells become polarized and cytoplasmic bridges have formed and are signified by the grey color. H) The infection thread are initiated. I) The infection thread begin to grow down the root hair. The nucleus moves through the root hair at the front of the thread. J) The infection thread fuses with the epidermal cell wall and the rhizobia grows into the intracellular space between the epidermal cell and the cortical cell. K) Continued growth of the infection thread and the outer cortical cell. L) Enlarged view of the root hair in I. The curl is unrolled and shows that the bacteria in the infection thread are outside fo the root hair. The plant cell wall is a black line an the plant cell membrane is a dashed line. Microtubules can be found in between the nucleus and the infection thread tip (blue). FromDaniel J. Gage |}
Nitrogen Fixation
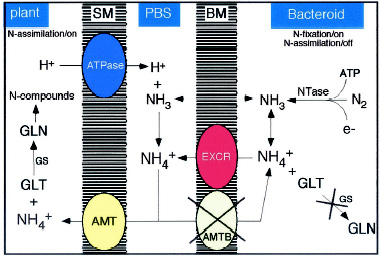
NH4+ flux and assimilation inside cells invaded with rhizobia. NH3 produce by nitrogenase from the bacteria diffuses through the BT membrane (labelled BM) and is converted into NH4+. NH4+ cannot be assimilated into glutamate (GLT) and glutamine (GLN) through glutamine synthetase (GS) and is therefore secreted most likely by an active mechanism (EXCR) in to the peribacteroid space (PBS) where an ATPase on the symbiosome membrane (SM) pumping protons keeps a high NH4+/NH3 ratio, and a transport system for NH4+ uptake (AMT) moves NH4+ to the plant cytoplasm where it is assimilated. From Patriarca et al. Read the full article for more information on bacterial NH4+ metabolism in Rhizobium-plant symbiosis.
Chemical Disruption
The estrogen receptor (ER) has been phylogenecally shown to be the ancestral receptor "from which all other steroid receptors have evolved" (Fox 2004). ERs bind endogenous estrogens as well as phytoestrogens, a molecule to which nodulation D protein (NodD) receptors in Rhizobium also bind. Many of the chemicals, synthetic and natural, that disrupt endocrine signaling in vertebrates disrupt phytoestrogen-NodD receptor signaling in rhizobia and other soil bacteria as well. Some of these chemicals include phytoestrogens, fungal chemicals, insecticides, herbicides, plasticizers, polyaromatic hydrocarbons (PAHs), and polychlorinated biphenyls (PCBs). If organochlorine pesticides and pollutants in high concentrations disrupt endocrine signaling in such a way that feminizes fish, reduces phallus sizes of reptiles, decreases mating abilities in mammals, reduces fertility and viability in different species, etc., what could it being doing to the Rhizobium-plant mutual system? The effects of these chemicals on the ecosystem might in fact be more tramatic than what was once thought. More research is being done on the evolution of chemical communication that occurs among plants, bacteria such as Rhizobium, and fungi to learn about the complex systems (Fox 2004).
References
Kaneko, Takakazu, Yasukazu Nakamura, Shusei Sato, Kiwamu Minamisawa, Toshiki Uchiumi, Shigemi Sasamoto, Akiko Watanabe, Kumi Idesawa, Mayumi Iriguchi, et al. "Complete genomic sequence of nitrogen-fixing symbiotic bacterium Bradyrhizobium japonicum USDA110."Universal Academy Press, Inc. 2001.