Roseobacter denitrificans: Difference between revisions
| Line 40: | Line 40: | ||
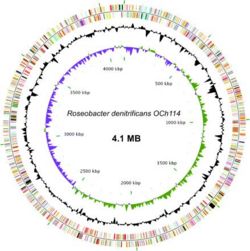
The genome sequence of ''Roseobacter denitrificans'' sp OCh 114 was completed in 2006 by a team in Arizona State University as the first AAP sequenced. (12) ''R. denitrificans'' contains a circular chromosome of 4,133,097 bp and four plasmids. Nearly half of the genes in the largest plasmid (pTB1) share a high degree of similarity to genes in plasmid pSD25 from ''Ruegeria'' sp. isolate PR1b and the Ti plasmid from ''Agrobacterium tumefaciens'' strain C58. (8) The conservation of many DNA integration and transfer proteins, plasmid replication proteins, and inverted repeats imply that R. denitrificans plasmid pTB1 may also promote lateral gene transfer. This possibility is especially interesting considering the propensity of R. denitrificans to associate with higher organisms. The genome of ''R. denitrificans'' additionally lacks genes that code for known photosynthetic carbon fixation pathways, with most notably missing the genes for Calvin cycle enzymes ribulose bisphosphate carboxylase (RuBisCO) and phosphoribukolkinase. (2) | The genome sequence of ''Roseobacter denitrificans'' sp OCh 114 was completed in 2006 by a team in Arizona State University as the first AAP sequenced. (12) ''R. denitrificans'' contains a circular chromosome of 4,133,097 bp and four plasmids. Nearly half of the genes in the largest plasmid (pTB1) share a high degree of similarity to genes in plasmid pSD25 from ''Ruegeria'' sp. isolate PR1b and the Ti plasmid from ''Agrobacterium tumefaciens'' strain C58. (8) The conservation of many DNA integration and transfer proteins, plasmid replication proteins, and inverted repeats imply that R. denitrificans plasmid pTB1 may also promote lateral gene transfer. This possibility is especially interesting considering the propensity of R. denitrificans to associate with higher organisms. The genome of ''R. denitrificans'' additionally lacks genes that code for known photosynthetic carbon fixation pathways, with most notably missing the genes for Calvin cycle enzymes ribulose bisphosphate carboxylase (RuBisCO) and phosphoribukolkinase. (2) | ||
Nearly 15% of the ''Roseobacter denitrificans'' genome is dedicated to the construction of transport proteins. Among these transporters are more than 120 genes coding for peptide and amino acid transporters(many branched chain), more than 110 genes coding | |||
for sugar and carbohydrate transporters, and more than 70 cation and iron transporters. These help the bacteria to acquire nitrogen other than fixing dinitrogen and allow R. denitrificans to adapt to the uptake of a variety of available carbon sources. (2) | |||
Revision as of 02:34, 29 August 2007
A Microbial Biorealm page on the genus Roseobacter denitrificans
Classification
Higher order taxa
Bacteria, Proteobacteria, Alphaproteobacteria, Rhodobacterales, Rhodobacteraceae, Roseobacter
Species
Roseobacter denitrificans sp. OCh114 (previously called Erythrobacter sp. OCh114)
Description and significance
Roseobacter denitrificans is a purple aerobic anyoxygenic phototrophic (AAP) bacterium that dwells free-living in lakes and ocean surface waters, soils and even near deep sea hydrothermal vents. (1,5) It can be isolated from the surfaces of green seaweeds of the coastal marine sediments in Australia, using proteinase K treatment followed by phenol extraction. (10,2) The first instance of its isolation however, was from the samples collected in aerobic marine environments: thalli of Enteromorpha linza, Porphyra sp., Sargussum horneri; beach sand; and the surface seawater from Aburatsubo Inlet in March and May 1978. (13) Members of the Roseobacter clade are widespread and abundant in such marine environments, having diverse metabolisms. The purple proteobacteria in particular, are the only known organisms to capture light energy to enhance growth requiring the presence of oxygen yet do not produce oxygen themselves. It grows photoautotrophically only at low oxygen levels, while at higher oxygen levels, the photosynthetic apparatus is down-regulated, resulting in chemotrophic growth using organic carbon. (2) The highly adaptive AAPs compose more than 10% of the microbial community in some euphotic upper ocean waters and are potentially major contributors to the fixation of the greenhouse gas CO2. The marine AAP species R. denitrificans grows not only photoheterotrophically in the presence of oxygen and light but also anaerobically in the dark using nitrate or trimethylamine N-oxide as an electron acceptor. It is the most studied AAP for this reason and is one of the main model organisms to study aerobic phototrophic bacteria. (2)
The importances of the bacterium's genome sequence are for the following reasons: 1) The evolutionary genesis of photosynthesis genes - it is a marine bacterium, and so may be representative of the globally huge population of aerobic phototrophic bacteria and help narrow their true evolutionary position by whole genome comparisons; (4,7) 2) Pathways of carbon dioxide fixation and production - in understanding the processes for its autotrophic CO2 production and the key genes involved in AAP CO2 fixation pathways. As the only aerobic phototrophic bacterium capable of anaerobic growth with use of nitrate as a terminal electron acceptor, it can also be used to facilitate subsequent studies of the effects of oxygen on photosynthetic and other metabolic processes; (5,6) 3) Light and oxygen signal transduction in gene expression - offering an opportunity, through bioinformatic approaches, to use the AAP's exaggerated responses to study of both light- and oxygen-responsive pathways and mechanisms that are difficult to study in other species. (3,4,11)
Genome structure
The genome sequence of Roseobacter denitrificans sp OCh 114 was completed in 2006 by a team in Arizona State University as the first AAP sequenced. (12) R. denitrificans contains a circular chromosome of 4,133,097 bp and four plasmids. Nearly half of the genes in the largest plasmid (pTB1) share a high degree of similarity to genes in plasmid pSD25 from Ruegeria sp. isolate PR1b and the Ti plasmid from Agrobacterium tumefaciens strain C58. (8) The conservation of many DNA integration and transfer proteins, plasmid replication proteins, and inverted repeats imply that R. denitrificans plasmid pTB1 may also promote lateral gene transfer. This possibility is especially interesting considering the propensity of R. denitrificans to associate with higher organisms. The genome of R. denitrificans additionally lacks genes that code for known photosynthetic carbon fixation pathways, with most notably missing the genes for Calvin cycle enzymes ribulose bisphosphate carboxylase (RuBisCO) and phosphoribukolkinase. (2)
Nearly 15% of the Roseobacter denitrificans genome is dedicated to the construction of transport proteins. Among these transporters are more than 120 genes coding for peptide and amino acid transporters(many branched chain), more than 110 genes coding for sugar and carbohydrate transporters, and more than 70 cation and iron transporters. These help the bacteria to acquire nitrogen other than fixing dinitrogen and allow R. denitrificans to adapt to the uptake of a variety of available carbon sources. (2)
The Complete Genome Sequence of Roseobacter denitrificans Reveals a Mixotrophic Rather than Photosynthetic Metabolism provides a possible explanation of how CO2 is fixated and produced using the complete genome sequence. "Based on the genome sequence of R. denitrificans, we suggest that they fix CO2 mixotrophically by using a sequestration pathway supplemented with additional CO2 provided by CO oxidation and heterotrophic respiration." (2)
Cell structure and metabolism
Describe any interesting features and/or cell structures; how it gains energy; what important molecules it produces.
The bacteria are gram-negative rods motile by means of polar or subpolar flagella, oxidase positive, and catalase positive, and produce a small amount of acids from glucose, fructose, xylose, and the other carbohydrates. The primary photosynthetic pigment of AAPBs is bacteriochlorophyll a. The amount of bacteriochlorophyll a found in Och 114 was 5.5 nmol/mg (dry weight). (13) This cellular bacteriochlorophyll a content is rather low, and the genes coding the enzymes synthesizing bacteriochlorophyll a may not be phenotype-expressed when limited by environmental conditions. (6)
They have a broad range of physiological properties and use a multitude of different carbon sources. Additional energy is gained by some organisms through oxidization of reduced sulfur compounds such as sulfite or thiosulfate, and, as stated above, some organisms possess bacteriochlorophyll a and are capable of aerobic photosynthesis; however, they are unable to grow autotrophically. (13,14)
Roseobacter denitrificans which lacks N2O reductase
Ecology
Describe any interactions with other organisms (included eukaryotes), contributions to the environment, effect on environment, etc.
Pathology
There are no known pathological effects of this bacterium on humans.
Application to Biotechnology
Does this organism produce any useful compounds or enzymes? What are they and how are they used?
Current Research
Enter summaries of the most recent research here--at least three required
References
1) Fleischman D and Kramer D (1998) Photosynthetic rhizobia. Biochim Biophys Acta 1364: 17-36.
10) http://www.roseobase.org/roseo/och114.html
11) http://genomes.tgen.org/rhodobacter.html
14) Sorokin, DY (1995) Sulfitobacter pontiacus gen. nov., sp. nov.—a new heterotrophic bacterium from the Black-Sea, specialized on sulfite oxidation. Microbiology 64: 295–305
Edited by George Huang student of Rachel Larsen