Yersinia: Difference between revisions
No edit summary |
No edit summary |
||
| Line 1: | Line 1: | ||
{| | {| | ||
| width="84" height="69" bgcolor="#FFDF95" | | | width="84" height="69" bgcolor="#FFDF95" | | ||
'''NCBI:<br />[http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Undef&id=629&lvl=3&keep=1&srchmode=1&unlock Taxonomy]<br /> Genome<br />'''''<font size="2">[http://www.ncbi.nlm.nih.gov/genomes/framik.cgi?db=Genome&gi=201 Y. pestis]</font>''[http://www.ncbi.nlm.nih.gov/genomes/framik.cgi?db=Genome&gi=201 <font size="2"> CO92</font><font size="2"></font>]<font size="2"><br />''[http://www.ncbi.nlm.nih.gov/genomes/framik.cgi?db=Genome&gi=250 Y. pestis ]''[http://www.ncbi.nlm.nih.gov/genomes/framik.cgi?db=Genome&gi=250 KIM''''''] </font> | '''NCBI:<br />[http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Undef&id=629&lvl=3&keep=1&srchmode=1&unlock Taxonomy]<br /> Genome<br />''' ''<font size="2">[http://www.ncbi.nlm.nih.gov/genomes/framik.cgi?db=Genome&gi=201 Y. pestis]</font>''[http://www.ncbi.nlm.nih.gov/genomes/framik.cgi?db=Genome&gi=201 <font size="2"> CO92</font><font size="2"></font>]<font size="2"><br />''[http://www.ncbi.nlm.nih.gov/genomes/framik.cgi?db=Genome&gi=250 Y. pestis ]''[http://www.ncbi.nlm.nih.gov/genomes/framik.cgi?db=Genome&gi=250 KIM''''''] </font> | ||
|} | |} | ||
[[Image:yersinia_1.jpg|thumb|350px|right|Image of ''Yersinia pestis.'' From [http://www.kvarkadabra.net/index.html?/biologija/teksti/biolosko_orozje.htm Kvarkadabra.]]] | [[Image:yersinia_1.jpg|thumb|350px|right|Image of ''Yersinia pestis.'' From [http://www.kvarkadabra.net/index.html?/biologija/teksti/biolosko_orozje.htm Kvarkadabra.]]] | ||
Revision as of 19:26, 12 June 2006
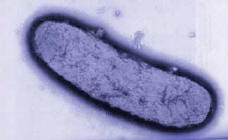
Classification
Higher order taxa:
Bacteria; Proteobacteria; Gammaproteobacteria; Enterobacteriales; Enterobacteriaceae
Species:
Yersinia pestis CO92, Y. pestis, Y. enterocolitica, Y. pseudotuberculosis
Description and Significance
Yersinia spp. are responsible for disease syndromes ranging from gastroenteritis to plague. Y. pestis is the cause of the plague and is actually catagorized into three subtypes or biovars; Antiqua, Medievalis, and Orientalis, each associated with a major pandemic. Y. pestis strand KIM belongs to biovar Mediaevalis while strand CO92 is in biovar Orientalis. Biovar Mediaevalis is thought to of descended from the bacteria that caused the second pandemic (the Black Death), while biovar Orientalis bacteria are responsible for the current pandemic (modern plague).
It is believed that Y. pestis is a clone that evolved from Y. pseudotuberculosis about 1.5 to 20 thousand years ago. This means that Y. pestis has evolved rapidly from being a pathogen widely found in the environment, able to infect mammalian intestines, to a blood-borne pathogen of mammals that can parasitize insects and has a limited capacity for survival outside a host.
Genome Structure
Y. pestis CO92 has a circular chromosome that is 4.65Mb in size with three plasmids pMT1 (96.2kb), pPCP1 (9.6kb), and pCD1 (70.3kb). The other Y. pestis strain sequenced, strain KIM, has the same three plasmids as well as a circular chromosome, but is 50kb smaller with 4.6Mb. When the two genomes were compared, a lot of similarity was found. The genomes share more than 95% of their sequence and 3,672 of 4,198 total ORFs match. Strain CO92 has one less rRNA operon than KIM.
Cell Structure and Metabolism
Y. pestis is rod shaped, gram negative, and non-motile yet has two distinct flagellar gene clusters; one set is incomplete and the other has a truncated FldH, which is a transcriptional activator for the flagellar genes.
Y. pestis uses aerobic respiration and anaerobic fermentation to produce and consume hydrogen gas for energy.
Ecology
Yersinia species are pathogens whose environments are not rich in minerals and grow at temperatures ranging from about 26oC to 37oC.
Pathology
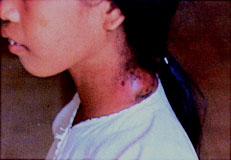
Y. pseudotuberculosis and Y. enterocolitica both infect the intestines of mammals through the fecal-oral route (contaminated food and water) and rarely is deadly. Y. pestis, on the other hand, is transmitted subcutaneously through a bite of an infected flea or rat (bubonic), but can also be transmitted by air (especially during pandemics of the disease). More specifically, fleas become infected after taking blood meals from septicemic animals and becoming infected themselves. Y. pestis grows in the midgut and eventually blocks the proventriculus, starving the flea for blood. The insects attempt to feed more often but end up giving back infected blood into the wound of the bite. The flea eventually dies, presumably from starvation and dehydration. On an interesting note, when the temperatures get higher fleas do not have their proventriculus blocked, and only those that are blocked can transmit the disease. When held at 30oC, fleas survive infections in an unblocked state, possibly leading at an explanation of why human bubonic plagues ended after the onset of warmer temperatures.
All pathogenic species of Yersinia contain the pCD1virulence plasmid. Y. pestis obtained two unique plasmids that encode a variety of virulence determinants. The pPCP1 plasmid encodes the plasminogen activator Pla, essential for virulence through the subcutaneous route. The pMT1 plasmid encodes murine toxin Ymt and the F1 capsular protein, which have been shown to play a role in the transmission of plague. Ymt, designated murine toxin because the protein is highly lethal for mice, is required for Y. pestis to survive in fleas. Ymt mutants, however, are as virulent as its parent in mice with plague.
Researchers have also discovered that at rising temperatures cells gradually lose their ability to bind Congo red (CR) along with their ability to cause disease in mice from peripheral routes of infection. This is because in the pgm (pigmentation) operon there are genes encoding the yersiniabactin (Ybt) siderophore-dependent iron transport system, required for virulence in mice subcutaneously, as well as genes for the Hms phenotype, which is required for cells to colonize and block the proventriculus.. A pgm deletion is therefore lethal to Y. pestis.
References
Parkhill et al. 2001. Genome sequence of Yersinia pestis, the causative agent of plague. Nature 413: 523-527. [Abstract]
Deng et al. 2002. Genome Sequence of Yersinia pestis KIM. Journal of Bacteriology 184 16: 4601-4611.
Achtman et al. 1999. Yersinia pestis, the cause of plague, is a recently emerged clone of Yersinia pseudotuberculosis. PNAS 96 24: 14043-14048.
Pepe, Jeffrey C. and Virginia L. Miller. 1993. Yersinia enterocoliticainvasion: A primary role in the initiation of infection. PNAS 90: 6473-6477.
Perry, Robert D. 2003. A Plague of Fleas - Survival and Transmission of Yersinia pestis. ASM News 69 7: 336-340.
