Pseudomonas aeruginosa: Difference between revisions
| Line 134: | Line 134: | ||
--Comparison between the ''P. aeruginosa'' PA01 strain and the more virulent ''P. aeruginosa'' PA14 was done to identity new virulence genes. First, shotgun genome sequencing was done on PA14 using 65,800 plasmids with 2-4 kb fragments of PA14 DNA. Then, a long-range PCR based method was implemented to determine if certain ''P. aeruginosa'' genomes are similar to PA01 or PA14 genomes. It was found that although PA14 gemone (6.5 Mbp) is somewhat larger than that of PA01 (6.3 Mbp), PA14 and PA01 genomes are very similar. There were 58 gene clusters from PA14 that were missing in PA01 and it was assumed that some of these genes are what make PA14 a lot more virulent than PA01. Microarray genomotyping of 18 diverse strains in the ''C. elegans'' model, however, showed that those 58 PA14 gene clusters did not correlate with these strains’ virulence. Thus a conclusion was drawn that the virulence in ''P. aeruginosa'' is both combinatorial and multifactorial and that the genes required for one strain to be pathogenic are not required for virulence in other strains (29). | --Comparison between the ''P. aeruginosa'' PA01 strain and the more virulent ''P. aeruginosa'' PA14 was done to identity new virulence genes. First, shotgun genome sequencing was done on PA14 using 65,800 plasmids with 2-4 kb fragments of PA14 DNA. Then, a long-range PCR based method was implemented to determine if certain ''P. aeruginosa'' genomes are similar to PA01 or PA14 genomes. It was found that although PA14 gemone (6.5 Mbp) is somewhat larger than that of PA01 (6.3 Mbp), PA14 and PA01 genomes are very similar. There were 58 gene clusters from PA14 that were missing in PA01 and it was assumed that some of these genes are what make PA14 a lot more virulent than PA01. Microarray genomotyping of 18 diverse strains in the ''C. elegans'' model, however, showed that those 58 PA14 gene clusters did not correlate with these strains’ virulence. Thus a conclusion was drawn that the virulence in ''P. aeruginosa'' is both combinatorial and multifactorial and that the genes required for one strain to be pathogenic are not required for virulence in other strains (29). | ||
Research also done in a microbiology lab at Loyola University Chicago has concluded that the Pseudomonas aeruginosa develops a greenish/yellow fluorescent pigment in nutrient broth and casein hydrolysis. After placing this fluorescent pigment under UV light, we observed a fluorescent blue/green pigment within the test tube. | [[Image: Team_6A_Fluoredit.JPG|frame|right| ''UV light test of fluorescent yellow/green pigment in nutrient broth'']] | ||
Research also done in a microbiology lab at Loyola University Chicago has concluded that the Pseudomonas aeruginosa develops a greenish/yellow fluorescent pigment in nutrient broth and casein hydrolysis. After placing this fluorescent pigment under UV light, we observed a fluorescent blue/green pigment within the test tube. | |||
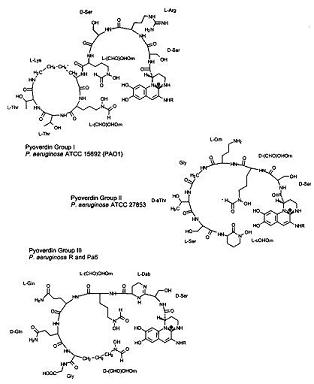
Under iron deficiency, a yellowish-green fluorescent pigment develops as a result of pyoverdins, a term named by Turfreijer for a group of compounds having a (1S)-5-amino-2,3-dihydro-8,9-dihydroxy-1H-pyrimido-[1,2a] chinolin-1-carboxylic acid chromophore. Figure 6A shows the different types of Pyoverdin groups that can be made on variations of their peptide chain. These pigment compounds only grow under iron limitation in a growth medium. [[Image: Pyoverdin.JPG|frame|left| ''Fig. 6A Thr'']]] By studying the structures of three pyroviderns in different P. aeruginosa strains we are able to quicky identify the bacteria | Under iron deficiency, a yellowish-green fluorescent pigment develops as a result of pyoverdins, a term named by Turfreijer for a group of compounds having a (1S)-5-amino-2,3-dihydro-8,9-dihydroxy-1H-pyrimido-[1,2a] chinolin-1-carboxylic acid chromophore. Figure 6A shows the different types of Pyoverdin groups that can be made on variations of their peptide chain. These pigment compounds only grow under iron limitation in a growth medium. [[Image: Pyoverdin.JPG|frame|left| ''Fig. 6A Thr'']]] By studying the structures of three pyroviderns in different P. aeruginosa strains we are able to quicky identify the bacteria | ||
Revision as of 14:56, 27 April 2010
A Microbial Biorealm page on the genus Pseudomonas aeruginosa
Classification
Higher order taxa
| Domain | Bacteria |
|---|---|
| Phylum | Proteobacteria |
| Class | Gamma proteobacteria |
| Order | Pseudomonadales |
| Family | Pseudomonadaceae |
| Genus | Pseudomonas |
| Species Group | Pseudomonas aeruginosa group |
Species
|
NCBI: Taxonomy |
| Genus species | Pseudomonas aeruginosa |
|---|

Description and significance
Pseudomonas aeruginosa is a gram-negative, rod-shaped, asporogenous, and monoflagellated bacterium that has an incredible nutritional versatility. It is about 1-5 µm in length and about 0.5-1.0 µm in breadth and is an obligate aerobe, which means it requires oxygen and uses aerobic respiration as its choice of metabolism. Due to its capability to synthesize arginine, P. aeruginosa can also proliferate in anaerobic conditions. This, then, makes P. aeruginosa a very ubiquitous microorganism, for it has been found in environments such as soil, water, humans, animals, plants, sewage, and hospitals (1). In all oligotropic aquatic ecosystems, which contain high-dissolved oxygen content but low plant nutrients throughout, P.aeruginosa is the predominant inhabitant and this clearly makes it the most abundant organism on earth (2).
P.aeruginosa is an opportunistic human pathogen. It is “opportunistic” because it seldom infects healthy individuals. Instead, it often colonizes immunocompromised patients, like those with cystic fibrosis, cancer, or AIDS (3). It is such a potent pathogen that firstly, it attacks up two thirds of the critically-ill hospitalized patients, and this usually portends more invasive diseases. Secondly, P.aeruginosa is the leading etiology for Gram-negative bacteria at most medical centers, carrying a 40-60% mortality rate. Thirdly, it complicates 90% of cystic fibrosis deaths; and lastly, it is always listed as one of the top three most frequent Gram-negative pathogens and is linked to the worst visual diseases (4). Furthermore, P.aeruginosa is a very important soil bacterium that is capable of breaking down polycyclic aromatic hydrocarbons and making rhamnolipids, quinolones, hydrogen cyanide, phenazines, and lectins (5). It also exhibits intrinsic resistance to a lot of different types of chemotherapeutic agents and antibiotics, making it a very hard pathogen to eliminate (1).
P. aeruginosa was first described as a distinct bacterial species at the end of the nineteenth century, after the development of sterile culture media by Pasteur. In 1882, the first scientific study on P. aeruginosa, entitled “On the blue and green coloration of bandages,” was published by a pharmacist named Carle Gessard. This study showed P. aeruginosa’s characteristic pigmentation: P. aeruginosa produced water-soluble pigments, which, on exposure to ultraviolet light, fluoresced blue-green light. This was later attributed to pyocyanine, a derivative of phenazine, and it also reflected the organism’s old names: Bacillus pyocyaneus, Bakterium aeruginosa, Pseudomonas polycolor, and Pseudomonas pyocyaneus (3). P. aeruginosa has many strains, including Pseudomonas aeruginosa strain PA01, Pseudomonas aeruginosa PA7, Pseudomonas aeruginosa strain UCBPP-PA14, and Pseudomonas aeruginosa strain 2192 (5). Most of these were isolated based on their distinctive grapelike odor of aminoacetophenone, pyocyanin production, and the colonies’ structure on agar media (6).
Genome structure
P. aeruginosa has the genome size of about 5.2 to 7 million base pairs (Mbp) with 65% Guanine + Cytosine content. It is a combination of variable accessory segments and a conserved core. The variable accessory genome is characterized by a set of genomic islands and islets from a primeval tRNA-integrated island type. The core genome consists of a low level of nucleotide divergence of 0.5% and a conserved synteny of genes, which means two or more genes, whether they are linked or not, are on the same chromosome (7).
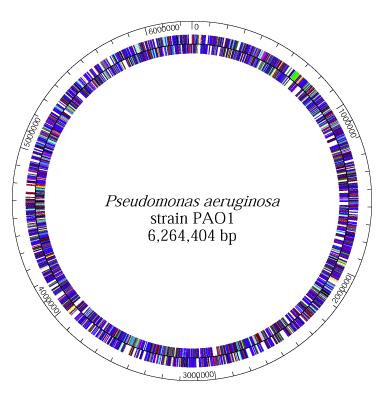
P. aeruginosa has a single and supercoiled circular chromosome in the cytoplasm (4). It also carries a lot of chromosome-mobilizing plasmids that are very significant to the organism’s lifestyle as a pathogen. The plasmids, TEM, OXA, and PSE, for instance, are encoded for betalactamase production, which is necessary for its resistance to antibiotics, thus allowing P. aeruginosa to be a formidable pathogen (8).
The two strains that have the complete genome sequence are Pseudomonas aeruginosa PA01 and Pseudomonas aeruginosa PA14 (9):
--In 2000, a group of volunteer "Pseudomonas scientists", including those from the Washington PathoGenesis Corportaion and the Department of Biology of the University of California, San Diego, worked under the Pseudomonas aeruginosa Community Annotation Project (PseudoCAP) to publish the complete genome sequence of Pseudomonas aeruginosa PA01. This was done because knowing the genomic sequence would provide new information about this bacterium as a pathogen and about its ecological versatility and genetic complexity. At 6,264,403 base pairs, its bacterial genome is the largest to ever be sequenced. It also contains 5,570 predicted open reading frames (ORFs), and thus it almost has the genetic complexity of simple eukaryotes, such as Saccharomyces cerevisiae. Using whole-genome-shotgun sampling, the complete 6.3 Mbp genome of Pseudomonas aeruginosa PA01 is very much similar to the P. aeruginosa’s physical map, with only one major exception, which is the inversion of about a quarter of the Pseudomonas aeruginosa PA01 genome. This inversion comes from the homologous recombination of the rrnA and rrnB loci, and earlier studies on genomic sequence inversions of ribosomal DNA loci in S. typhimurium and E. coli suggest that this inversion might have adaptive significance (10).
--The complete genome sequence of Pseudomonas aeruginosa PA14 is currently being done by Harvard Medical School scientists. The goal of this study is achieve a public data of Pseudomonas aeruginosa PA14 genome. The shotgun-sequencing phase of the project was finished in 2005, yielding 6.54 Mbp of PA14 sequence. It is currently being compared to the genome of Pseudomonas aeruginosa PA01 and preliminary results have shown that they are very similar but have several regions of marked differences, such as the insertion of the 107911bp in PA14, which is absent in PA01. Approximately, there is 96.3% of the DNA sequence of PAO1 is in PA14, and 92.4% of PA14 DNA sequence is in PA01 (11).
Cell structure and metabolism
Protein F--Since P. aeruginosa is a Gram-negative microbe, it has an outer membrane which contains Protein F (OprF). OprF functions as a porin, allowing certain molecules and ions to come into the cells, and as a structural protein, maintaining the bacterial cell shape. Because OprF provides P. aeruginosa outer membrane with an exclusion limit of 500 Da, it lowers the permeability of the outer membrane, a property that is desired because it would decrease the intake of harmful substances into the cell and give P. aeruginosa a high resistance to antibiotics (12).
Flagellum and Pili--P. aeruginosa uses its single and polar flagellum to move around and to display chemotaxis to useful molecules, like sugars. Its strains either have a-type or b-type of flagella, a classification that is based primarily on the size and antigenicity of the flagellin subunit. The flagellum is very important during the early stages of infection, for it can attach to and invade tissues of the hosts (13). Similarly to its flagellum, P. aeruginosa pili contribute greatly to its ability to adhere to mucosal surfaces and epithelial cells. Specifically, it is the pili’s tip that is responsible for the adherence to the host cell surface. P. aeruginosa have N-methyl-phenyl-alanine (NMePhe) or type IV pili (1). The pili are characterized as long polar filaments made up of homopolymers from the protein pilin, which is encoded by the pilA gene (4). Overall, P. aeruginosa flagellum and pili have similar functionality (for attachment) and structure (both are filamentous structures on the surface of the cell), and their motility is controlled by RpoN, especially during initial attachment to the human host and under low nutrient conditions (1).
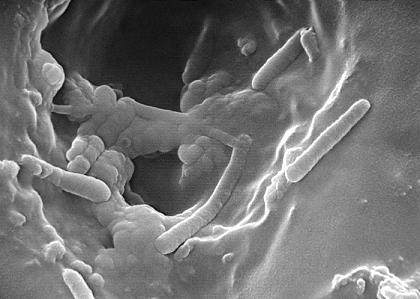
When infecting its host, P. aeruginosa is starved for iron because iron deprivation of an infecting pathogen is the key part in the humans’ innate defense mechanism. To overcome this challenge, P. aeruginosa synthesizes two siderophores: pyochelin and pyoverdin. P. aeruginosa then secrets these sideophores to the exterior of the cell, where they bind tightly to iron and bring the iron back into the cell. Additionally, P. aeruginosa can also use iron from enterobactin, a special siderophore produced by E. coli for iron transport, to satisfy its iron need (14).
P. aeruginosa is a facultative aerobe; its preferred metabolism is respiration. It gains energy by transferring electrons from glucose, a reduced substrate, to oxygen, the final electron acceptor (15). The breakdown of glucose requires it to oxidize to gluconate in the periplasm, then it will be brought inside the inner membrane by a specific energy-dependent gluconate uptake system. Once inside, gluconate is phosphorylated to 6-P-gluconate, which will enter the central metabolism to produce energy for the cell (16). When P. aeruginosa is in anaerobic conditions, however, P. aeruginosa uses nitrate as a terminal electron acceptor(17). Under oxidative-stress conditions, P. aeruginosa synthesizes Fe- or Mn- containing superoxide dismutase (SOD) enzymes, which catalyze the very reactive O- to H2O2 and O2. It also detoxifies H2O2 to O2 and H2O by using catalase (1).
Ecology
Since P. aeruginosa can live in both inanimate and human environments, it has been characterized as a “ubiquitous” microorganism. This versatility is made possible by a large number of enzymes that allow P. aeruginosa to use a diversity of substances as nutrients. Most impressively, P. aeruginosa can switch from growing on nonmucoid to mucoid environments, which comes with a large synthesis of alginate. In inanimate environment, P. aeruginosa is usually detected in water-reservoirs polluted by animals and humans, such as sewage and sinks inside and outside of hospitals. It is also found in swimming pools and whirlpools because the warm temperatures are favorable to its growth (3). Because it thrived in warm conditions, however, it was determined to be the culprit of the Hot Tub Rash, in which direct contact between the skin and the infected water from the tub will make the infected skin itchy and turn it a bumpy red color (19). In addition, P. aeruginosa is an opportunistic human pathogen that causes chronic infections in patients with cystic fibrosis and is the leading cause of death by Gram-negative bacteria (more under pathology) (3).
Although most P. aeruginosa-plant interactions are detrimental to the plant, a recent study has found a P. aeruginosa strain that actually supports plant growth. This characteristic, along with the fact that P. aeruginosa can degrade polycyclic aromatic hydrocarbons, suggests the future uses of P. aeruginosa for environmental detoxification of synthetic chemicals and pesticides and for industrial purposes (3).
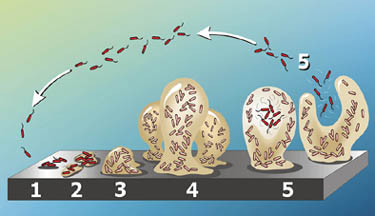
P. aeruginosa groups tend to form biofilms, which are complex bacterial communities that adhere to a variety of surfaces, including metals, plastics, medical implant materials, and tissue. Biofilms are characterized by “attached for survival” because once they are formed, they are very difficult to destroy. Depending on their locations, biofilms can either be beneficial and detrimental to the environment. For instance, the biofilms found on rocks and pebbles underwater of lakes and ponds are an important food source for many aquatic organisms. On the contrary, those that developed on the interiors of water pipes might cause clogging and corrosions (19) (20).
Pathology

P. aeruginosa rarely causes disease in healthy humans. It is usually linked with patients whose immune system is compromised by diseases or trauma. It gains access to these patients’ tissues through the burns, for the burn victims, or through an underlying disease, like cystic fibrosis. First, P. aeruginosa adheres to tissue surfaces using its flagellum, pili, and exo-S; then, it replicates to create infectious critical mass; and lastly, it makes tissue damage using its virulence factors (21). Since the powerful exotoxins and endotoxins released by P. aeruginosa during bacteremias continue to infect the host even after P. aeruginosa has been killed off by antibiotics, acute diseases caused by P. aeruginosa tend to be chronic and life-threatening. Furthermore, with the exception of the cystic fibrosis strain, most P. aeruginosa strains that attack compromised patients tend to be nonmucoid (2). And even though a small amount of patients infected by P. aeruginosa developed severe sepsis with lesions with black centers, most patients exhibited no obvious pathological effects of the colonization (22).
Cystic fibrosis (CF) is the most common autosomal recessive disorder in Caucasians. With a mutation on chromosome 7, a CF lung cannot transport chloride (Cl-), sodium (Na+), and water from the basolateral to the secretory epithelia. This disruption in the salt and water balance in the cell results in the production of a thick mucus, which becomes the ideal home for potential pathogens. P. aeruginosa attacks CF patients via airway and once it is in, it uses its flagellum to go to the hypoxic zone, an oxygen-depleted environment. At this location, P. aeruginosa undergoes a transition from an aerobic to an anaerobic microbe and starts forming biofilms anaerobically. Once this is formed, the P. aeruginosa in this community can sense their population via quorum sensing, where they secret low molecular weight pheromones that enable them to communicate with each other (23). This gives them the ability to resist many defenses, including anti-Pseudomonas antibiotics such as ticarcillin, ceftazidime, tobramycin, and ciprofloxacin, because once the bacteria sense that their outer layer of biofilm is being destroyed, the inner layers will grow stronger to reestablish the community (24). P. aeruginosa is also resistant to many antibiotics and chemotherapeutic agents due to their intrinsic resistance. This is caused by the low permeability to antibiotics of the outer membrane and by the production of β-lactamases against multidrug efflux pumps and β-lactam antibiotics (22).
P. aeruginosa communicates with other cells through quorum-sensing. This form of communication allows the cells to regulate gene production which results in control of certain cell functions. One of the enzymes responsible for quorum sensing is tyrosine phosphatase (TpbA). This enzyme relays extracellular quorum sensing signals to polysaccharide production and biofilm formation outside the cells (32). P. aeruginosa attaches to surfaces by way of biofilm production. Quorum-sensing can be a drug target to cure infections caused by P. aeruginosa. Quorum-quenching is used to blocks the signaling mechanism of quorum-sensing and prevents biofilm formation in P. aeruginosa. Yi-Hu Dong and his colleagues were able to prevent biofilm formation in mice under laboratory conditions (33).
P. aeruginosa secrets many virulent factors to colonize the cells of its host. For example, exotoxin A, the most toxic protein produced by P. aeruginosa, catalyzes the ADP-ribosylation to form ADP-ribosyl-EF-2, which inhibits the protein synthesis of the host’s cells. Moreover, elastase, an extracellular zinc protease, attacks eukaryotic proteins such as collagen and elastin and destroys the structural proteins of the cell. It also breaks down human immunoglobin and serum alpha proteins (1).
Furthermore, P. aeruginosa infects animals. In an experiment, intravenous injection of virulent P. aeruginosa was injected into mice and these animals usually died within 24-48 hours. When a smaller dose was injected, characteristic signs of infection such as weight loss, focal lesions in liver, spleen, and kidneys, followed by death within 3-10 days, would take place. P. aeruginosa has also been found to cause outbreaks of pneumonia in guinea pigs, and although it also attacks plants, not a lot of research has been done in this area (22).
Application to Biotechnology
P. aeruginosa, as well as many other Pseudomonas, can degrade aromatic hydrocarbons such as methylbenzenes, which are the by-products of petroleum industries and are commonly used as solvents for enamels and paints as well as in the production of drugs and chemicals. Methylbenzenes are considered as environmental contaminants that are present in the atmosphere, underground and soils, and in surface water (25). P. aeruginosa can break down toluene, the simplest form of methylbenzene. P. aeruginosa degrades toluene through the oxidation of the methyl group to aldehyde, alcohol, and an acid, which is then converted to catechol. Hence, P. aeruginosa can be used in pollution control (26).
Current Research
Effect of Spaceflight on Microbial Gene Expression and Virulence (Microbe)
--The National Aeronautics and Space Administration (NASA) and the Biodesign Institute at Arizona State University are currently carrying out a research project called the Microbe Experiment. In this experiment, three microbial pathogens Pseudomonas aeruginosa, Salmonella typhimurium, and Candida albicans are being brought into space to see how their genetic responses and virulence change. These three microbes have been viewed as potential threat to the health of the astronauts, for P. aeruginosa had contaminated the spacecraft’s water system and infected a crew member during the Apollo era. Thus, understanding their adaptation and virulence in microgravity will give scientists more information about the crew’s space environment and better prepare the astronauts for future space explorations. The microbes were placed inside self-contained culture chambers and upon landing back on earth, one thirds of the sample will be used for virulence studies while the remaining will be kept frozen at -80oC. Because this is an ongoing research project, there have not been any results but NASA scientists are very hopeful that this study will lead to novel discoveries of vaccines against these microbes here on Earth and during spaceflight (27).
The Combination of PCR and Serology Increases the Diagnosis of Pseudomonas aeruginosa Colonization/Infection in Cystic Fibrosis
--Microbiological culturing methods are often used for the early diagnosis of P. aeruginosa infection in cystic fibrosis (CF) patients. These methods, however, have some disadvantages because P. aeruginosa might not be detected since initial infection is usually in low density. It was then proposed that serology and polymerase chain reaction (PCR) might be better techniques in detecting the early stage of P. aeruginosa infection in children with CF. The experiment was carried out by collecting sputum and serum from 87 CF children with a mean age of 9.7 years. Then, 1) PCR was performed on the sputum, targeting P. aeruginosa algD GDP mannose dehydrogenase gene. 2) Serology was done against P. aeruginosa antigens: exotoxin A, elastase, and alkaline protease. 3) A combination of PCR and serology was done. When looking at the results, using the PCR or serology method alone did not yield statistically significant difference from the microbiological culturing methods. The combination of PCR and serology, however, identified a lot more patients than any of the two methods alone. Hence, a combination method that includes PCR will be an accurate technique to use in early diagnosis of P. aeruginosa colonization in CF patients (28).
Genomic analysis reveals that Pseudomonas aeruginosa virulence is combinatorial
--Comparison between the P. aeruginosa PA01 strain and the more virulent P. aeruginosa PA14 was done to identity new virulence genes. First, shotgun genome sequencing was done on PA14 using 65,800 plasmids with 2-4 kb fragments of PA14 DNA. Then, a long-range PCR based method was implemented to determine if certain P. aeruginosa genomes are similar to PA01 or PA14 genomes. It was found that although PA14 gemone (6.5 Mbp) is somewhat larger than that of PA01 (6.3 Mbp), PA14 and PA01 genomes are very similar. There were 58 gene clusters from PA14 that were missing in PA01 and it was assumed that some of these genes are what make PA14 a lot more virulent than PA01. Microarray genomotyping of 18 diverse strains in the C. elegans model, however, showed that those 58 PA14 gene clusters did not correlate with these strains’ virulence. Thus a conclusion was drawn that the virulence in P. aeruginosa is both combinatorial and multifactorial and that the genes required for one strain to be pathogenic are not required for virulence in other strains (29).
Research also done in a microbiology lab at Loyola University Chicago has concluded that the Pseudomonas aeruginosa develops a greenish/yellow fluorescent pigment in nutrient broth and casein hydrolysis. After placing this fluorescent pigment under UV light, we observed a fluorescent blue/green pigment within the test tube.
Under iron deficiency, a yellowish-green fluorescent pigment develops as a result of pyoverdins, a term named by Turfreijer for a group of compounds having a (1S)-5-amino-2,3-dihydro-8,9-dihydroxy-1H-pyrimido-[1,2a] chinolin-1-carboxylic acid chromophore. Figure 6A shows the different types of Pyoverdin groups that can be made on variations of their peptide chain. These pigment compounds only grow under iron limitation in a growth medium.
] By studying the structures of three pyroviderns in different P. aeruginosa strains we are able to quicky identify the bacteria
References
1) Lederberg, Joshua et al. Pseudomonas. Encyclopedia of Microbiology. Second Edition. Volume 3. San Diego, 2000. p. 876-891.
2) Costerton, W., and Anwar, H. Pseudomonas aeruginosa: The Microbe and Pathogen. Pseudomonas aeruginosa Infections and Treatment. 1994. p.1-17.
3) Botzenhardt, K., and Doring, G. Ecology and epidemiology of Pseudomonas aeruginosa. Pseudomonas aeruginosa as an Opportunistic Pathogen. 1993. p. 1-7.
4) Fick, R. Pseudomonas aeruginosa—the Microbial Hyena and Its Role in Disease: An Introduciton. Pseudomonas aeruginosa: The Opportunist. 1993. p. 1-6.
5) National Center for Biotechnology Information site
6) Gilardi, G. Cultural and Biochemical Aspects for Identification of Glucose-Nonfermenting Gram-Negative Rods. Nonfermenting Gram-Negative Rods. 1985. p.17-24.
8) Craig, W., and Ebert, S. Antimirobial Therapy in Pseudomonas aeruginosa Infections. Pseudomonas aeruginosa Infections and Treatment. 1994. p. 470-491.
9) Pseudomonas Genome Database
11) P. aeruginosa PA14 Genomic Sequencing Project
12) Stanisich, V., and Richmond, M. Gene Transfer in the Genus Pseudomonas. Genetics and Biochemistry of Pseudomonas. 1975. p. 170-175.
13) Delden, C. Virulence Fators in Pseudomonas aeruginosa. Pseudomonas. 2004. p. 1-7.
14) Cox, C. Iron and the Virulence of Pseudomonas aeruginosa. Pseudomonas aeruginosa: The Opportunist. 1993. p. 41-45.
16) Rojo, F., and Dinamarca, A. Catabolite Repression and Physiological Control. Pseudomonas. 2004. p. 365-366.
17) Valls, M., Cases, I., and Lorenzo, V. Transcription Mediated by rpoN-Dependent Promoters. Pseudomonas. 2004. p. 398-302.
18) Center for Diseases Control and Prevention
19) Center for Biofilm Engineering
20) Brown, M., and Simith, A. Antimicrobial Agents and Biofilms. Medical Implications of Biofilms. 2003. p. 36-38.
21) Irvin, Randall. Attachment and Colonization of Pseudomonas aerugionsa: Role of the Surface Structures. 'Pseudomonas aeruginosa as an Opportunistic Pathogen. 1993. p. 19-36.
22) Lowbury, E. Biological Importance of Pseudomonas aeruginosa: Medical Aspects. Genetics and Biochemistry of Pseudomonas. 1975. p. 37-43.
23) Holden, M., and Williams, P. “Quorum Sensing”. Encyclopedia of Life Sciences. 2001.
24) Smiley, A., and Hassett, D. Pseudomonas aeruginosa Biofilm Infections in Cystic Fibrosis. Biofilms, Infection, and Antimicrobial Therapy. 2006. p. 155-158.
25) Pieper, D., Stadler-Fritzche, K., Scholomann, M., and Knackmuss, H. Metabolism of 2-Chloro-4-Methylphenoxyacetate by Alcaligenes eutrophus JMP 134: Implications for the Degradation of Chloro- and Methyl-Substituted Aromatics via ortho Cleavage. Pseudomonas Molecular Biology and Biotechnology. 1992. p. 268-272.
26) Johnson, G., and Olsen, R. “Multiple Pathways for Toluene Degradation in Burkholderia sp. Strain JS150.” Applied and Environmental Microbiology. 1997. Volume 63. p. 4047-4052. [1]
27) The National Aeronautics and Space Administration
29) Lee, D., Urbach, J., Wu, G., Liberati, N., Feinbaum, R., Miyata, S., Diggins, L., He, J., Saucier, M., Déziel, E., Friedman, L., Li, L., Grills, G., Montgomery, K., Kucherlapati, R., Rahme, L., and Ausubel, F. “Genomic analysis reveals that Pseudomonas aeruginosa virulence is Combinatorial.” Genome Biology. 2007. Volume 7. [2]
30) Edited by students: Vivek Brahmbhatt and Varun Garg of M Glogowski at Loyola University. http://web.ebscohost.com/ehost/pdfviewer/pdfviewer?vid=4&hid=104&sid=0029bc6b-e73c-4b8c-8882-42c0b9d6cedf%40sessionmgr112.
31) Malone, Jacob G., Tina Jaeger, and Christian Spangler. "YfiBNR Mediates Cyclic Di-GMP Dependent Small Colony Variant Formation and Persistence in Pseudomonas Aeruginosa." PLoS Pathogens 7.3 (2010): 1-17. Academic Search Premiere. Web. 24 Apr. 2010.
Written by Chelsea Dao, a student of Rachel Larsen
Edited by KLB