Gemmatimonas aurantiaca: Difference between revisions
No edit summary |
|||
| Line 18: | Line 18: | ||
==Genome structure== | ==Genome structure== | ||
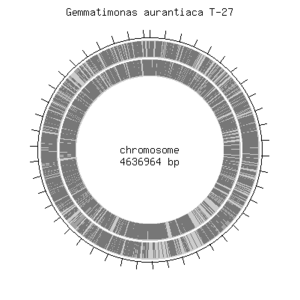
<i>Gemmatimonas aurantiaca T-27T</i> has a circular chromosome that consists of 4,636,964 base pairs of which majority, 64.28%, includes G/C pair. <i>G. aurantiaca</i> contains about 3,935 open reading frames, 48 tRNA genes and single rRNA operon were predicted. It also contains essential genes that were previously identified in model organisms such as <i>Escherichia coli</i> and <i>Bacillus subtilis</i>. This shows that their basic cellular system is not much different from other known microbes. <i>G. aurantiaca T-27T</> can grow both under aerobic and anaerobic conditions. Furthermore, <i>G. auratiaca T-27T</i> genome encoded significantly large numbers of signal transduction components, sigma factors and transporters. These genomic features would provide an insight into the life style of <i>G. aurantiaca</i>, and facilitate isolation of not-yet-cultivated Gemmatimonadetes species.<br> | |||
[[File:Genome_gemmatimonas.png|thumb|Genome structure of <i>G. aurantiaca</i> [Genome Information Broker]]] | [[File:Genome_gemmatimonas.png|thumb|Genome structure of <i>G. aurantiaca</i> [Genome Information Broker]]] | ||
Revision as of 14:16, 3 February 2012
A Microbial Biorealm page on the genus Gemmatimonas aurantiaca
Classification
Higher order taxa
Bacteria; Gemmatimonadetes; Gemmatimonadales; Gemmatimonadaceae; Gemmatimonas.
Species
Gemmatimonas aurantiaca T-27
Description and significance
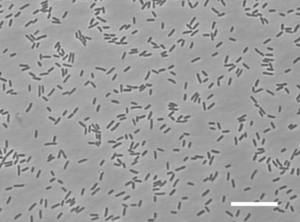
G. aurantiaca T27 is an aerobic rod-shaped Gram-negative bacterium that is non-sporulating. It is motile by the use of flagella and it was isolated by anaerobic-aerobic sequential batch reactor under enhanced biological phosphorus removal conditions for wastewater treatment. G. aurantiaca T27 is a very slow growing bacterium and is a polyphosphate-accummulating bacteria.
Genome structure
Gemmatimonas aurantiaca T-27T has a circular chromosome that consists of 4,636,964 base pairs of which majority, 64.28%, includes G/C pair. G. aurantiaca contains about 3,935 open reading frames, 48 tRNA genes and single rRNA operon were predicted. It also contains essential genes that were previously identified in model organisms such as Escherichia coli and Bacillus subtilis. This shows that their basic cellular system is not much different from other known microbes. G. aurantiaca T-27T</> can grow both under aerobic and anaerobic conditions. Furthermore, G. auratiaca T-27T genome encoded significantly large numbers of signal transduction components, sigma factors and transporters. These genomic features would provide an insight into the life style of G. aurantiaca, and facilitate isolation of not-yet-cultivated Gemmatimonadetes species.
Cell structure and metabolism
Interesting features of cell structure; how it gains energy; what important molecules it produces.
Ecology
Habitat; symbiosis; contributions to the environment.
Pathology
How does this organism cause disease? Human, animal, plant hosts? Virulence factors, as well as patient symptoms.
Current Research
Enter summaries of the most recent research here--at least three required
Cool Factor
Describe something you fing "cool" about this microbe.
References
Gemmatimonas aurantiaca T-27." Genome Information Broker. Web.
Zhang, Hui, Yuji Sekiguchi, Satoshi Hanada, Philip Hugenholtz, Hongik Kim, Yoichi Kamagata, and Kazunori Nakamura. "Gemmatimonas Aurantiaca Gen. Nov., Sp. Nov., a Gram-negative, Aerobic, Polyphosphate-accumulating Micro-organism, the First Cultured Representative of the New Bacterial Phylum Gemmatimonadetes Phyl. Nov." International Journal of Systematic and Evolutionary Microbiology.
Edited by student of Iris Keren: NEUS2012 Amal Mohamed, Soonji Kim, Jaclyn Egitto