Shewanella woodyi NEU Spring 2012: Difference between revisions
| Line 32: | Line 32: | ||
==Pathology== | ==Pathology== | ||
While other species of <i> Shewanella</i> are known pathogens (ie. <i>Shewanella marisflavi</i> isolate AP629 is pathogenic to sea cucumber), <i>S. woodyi</i> has not proven to be a pathogenic organism. [8] | |||
==Current Research== | ==Current Research== | ||
Revision as of 01:51, 17 February 2012
A Microbial Biorealm page on the genus Shewanella woodyi NEU Spring 2012
Classification
Bacteria; Proteobacteria; Gammaproteobacteria; Alteromonadales; Shewanellaceae; Shewanella; woodyi [3]
Description and significance
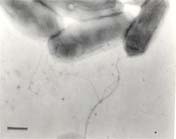
Shewanella woodyi is a gram negative bacilli shaped bacterium between 1 to 2 micrometers in length [1,4]. It is entirely marine and can be found in deep-sea environments therefore making it barophilic (tolerant to high pressure) [1,2]. Although other species within the Shewanella genus are psychrophilic, S. woodyi is instead mesophilic growing at an optimal temperature around 25°C [1,2]. Each S. woodyi bacterial cell possesses a single polar flagellum making it motile, as seen in Figure 1 [1]. This bacterium is also luminescent, meaning it can emit light[1].
It is well known that the ocean acts as a sink for harmful compounds from both terrestrial and aquatic environments. S. woodyi is significant in that it helps reduce the levels of select toxic compounds that collect in the sediment on the ocean floor [2,6]. Additionally, this bacterium also plays a role in carbon and nitrogen cycling through its anaerobic capabilities [2,6,7].
Genome structure
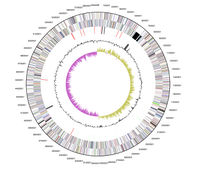
S. woodyi has a circular DNA chromosome of 5,935,403 bases, of which 86.02% are coding bases. This chromosome has a G+C content of 43.7%. The entire chromosome can be seen in Figure 2. These base pairs code for 5,085 genes in total, with 96.81%, or 4,923 genes, coding for proteins. The remaining 3.19%, or 162 genes, code for RNA.
Cell structure and metabolism
Shewanella woodyi are gram negative, luminescent, nonsporulating rods (0.4-1.0 by 1.4-2.0 pm) with an unsheathed flagellum as was mentioned above. The molar percent of G and C content of its DNA is 39 mol%. For this bacteria's metabolism, it is catalase and oxidase positive and most strains produce amylase. All strains do produce gelatinase but are unable to make lipase, chtinase, and agarase. They are able to metabolize D-galactose, cellobiose, D-glucuronic acid, acetate, a-ketoglutarate,propionate, succinate, L-alanine, L-threonine, L-leucine, L-serine, and putrescine. Oxygen and nitrite is used by these cells as electron acceptors for their metabolic processes.[1]
Ecology
Originally found in squid ink, ocean floor sediment, and among detritus from the Alboran Sea, this bacterium can be found in a wide range of areas [1]. It thrives at 25°C in a deep-sea environment where it can use detritus and other compounds present for energy [1,2,7]. Research in the past few years has suggested that these bacteria can use almost any electron acceptor for anaerobic respiration [7]. The only criteria seems to be that the electron acceptor must be more electronegative than sulfate [7].
High pressure and cold temperatures are two major factors that S. woodyi must overcome in its environment. Usually under these conditions, cell membranes become very rigid and stiff. S. woodyi avoids this problem by producing large amounts of polyunsaturated fatty acids in order to create a very lipid rich membrane [2,5]. This characteristic allows for a more fluid membrane even under high pressures and lower temperatures.
Many of these characteristics that S. woodyi have acquired to live in their environment are a result of horizontal gene transfer from other species [6]. For example, S. woodyi inherited its Na+/H+ antiporter pump from other bacteria that were already expressing that particular gene [6]. As a result, this bacteria is able to regulate the concentration of sodium in the cell in order to live in high salinity waters.
Pathology
While other species of Shewanella are known pathogens (ie. Shewanella marisflavi isolate AP629 is pathogenic to sea cucumber), S. woodyi has not proven to be a pathogenic organism. [8]
Current Research
http://www.decodedscience.com/nitric-oxide-a-newly-discovered-potential-target-for-biofilm-control/9730 http://www.sciencedirect.com/science/article/pii/S0167701211001308 http://www.biomedcentral.com/1471-2164/12/S1/S3
Cool Factor
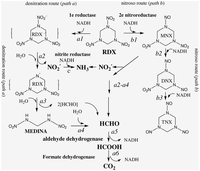
As previously mentioned, S. woodyi plays a role in alleviating the environment of toxic compounds. There are several organisms which also complete similar tasks and remove harmful components from the air or soil, however, not many are capable of degrading compounds such as RDX (hexahydro-1,3,5-trinitro-1,3,5-triazine) along with other explosives [6]. S. woodyi contains cytochrome c genes and nitroreductases which allow for the break down of RDX into nitric oxide and carbon dioxide [6]. This pathway is shown on the right side of the Figure 2 [6]. Although Shewanella halifaxensis and Shewanella sediminis are more efficient at breaking down RDX than S. woodyi and can use both degradation pathways shown in Figure 2, S. woodyi is still capable of this activity making it unique from many other microorganisms [6]. This factor also makes it even more important to the countless animals that depend on non-polluted waters for better survival. Recent research has suggested that in the presence of explosive compounds, such as RDX, bacteria in the Shewanella genus, including S. woodyi, actually thrive [6].
References
[1] Makemson, J. C., Fulayfil, N. R., Landry, W., Van Ert, L. M., Wimpee, C. F., Widder, E. A. & Case, J. F. (1997). Shewanella woodyi sp. nov., an exclusively respiratory luminous bacterium isolated from the Alboran Sea. Int J Syst Bacteriol 47, 1034-1039. [2] Kato, C., and Nogi, Y. (2001). Correlation between phylogenetic structure and function: examples from deep-sea Shewanella. FEMS Microbiology Ecology 35, 223-230. [3] "Shewanella Woodyi MS32, ATCC 51908." Integrated Microbial Genomes. U.S. Department of Energy Joint Genome Institute. Web. 02 Feb. 2012. [4] http://www.ncbi.nlm.nih.gov/sites/entrez?db=bioproject&cmd=ShowDetailView&TermToSearch=17455 [5] Wang, F., Xiao, X., Ou, H., Gai, Y., and Wang, F. (2009). Role and regulation of fatty acid biosynthesis in the response of S. pizotolerans WP3 to different temperatures and pressures. Journal of Bacteriology 191, 2574-2584. [6] Zhao, J., Deng, Y., Manno, D., and Hawari, J. (2010). Genomic evolution for a cold marine lifestyle and in-situ explosive biodegration. PLoS ONE 5, online. [7] Nealson, K.H., and Scott, J. Ecophysiology of the Genus Shewanella. (2006). Prokaryotes 6, 1133-1151.