Dehalobacter: Difference between revisions
| Line 38: | Line 38: | ||
===Reductive dehalogenation=== | ===Reductive dehalogenation=== | ||
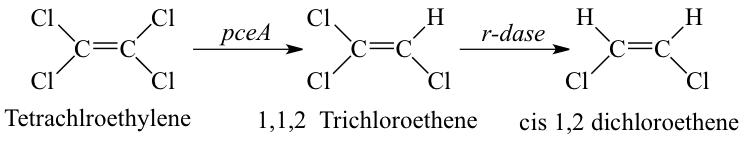
[[Image:Reductive_dehalogenation.png|frame|left|Metabolic pathway associated with organohalide respiration of Tetrachloroethylene mediated by Dehalobacter spp. resulting in incomplete reductive dechlorination to cis-1,2 dichlorethene (an environmentally unacceptable endpoint). From [Source, Hollinger et al., Arch Microbiol, 1998.]]] Reductive dehalogenation is the reduction of halogenated aliphatic and aromatic compounds using hydrogen as an electron donor. Microbially mediated reductive dehalogenation linked to growth is referred to as organohalide respiration. In addition to Dehalobacter restrictus, numerous microbial genera mediate organohalide respiration (e.g. Dehalococcoides, Dehalogimonas, etc.). When a chlorinated molecule is used as electron acceptor for organohalide respiration, and it is thus termed as microbially mediated reductive dechlorination. In this process, the chlorine atom on a chlorinated molecule is replaced by a hydrogen atom using a reductive dehalogenase enzyme (rdase) like pceA. | |||
Dehalobacter restrictus accepts chlorinated solvents, PCE and TCE, as electron acceptors. However, reductive dechlorination is incomplete and stops at cis-1,2 dichlorethene (cDCE). | |||
Dehalobacter Strain Known electron acceptors End Products Source | |||
PER K23 PCE, TCE cDCE (5) | |||
TCA1 1,1,1 TCA, 1,2 DCA Chloroethane (9) | |||
MS CF, 1,1,1 TCA, 1,2DCA DCM (10) | |||
Mixed populations of Dehalobacter spp. have also been shown to mediate reductive dehalogenation of 1,2 dichlorobenzene, monochlorobenzene, polychlorinated biphenyl and other recalcitrant chlorinated compounds. | |||
===Fermentation of Dichloromethane=== | |||
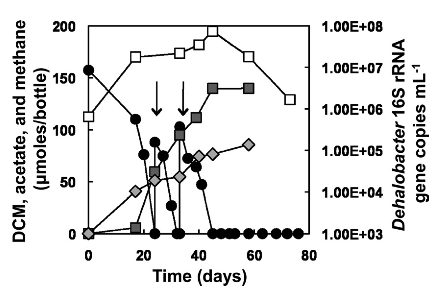
[[Image:Fermentation_of_dichloromethane.png|frame|below|Fermentative dehalogenation of DCM and a consequent increase in 16S rRNA gene copies of Dehalobacter gene copies. From [Justicia-Leon S.D. et al., 2012, AEM]]] Several different strains of Dehalobacter spp. have been known to degrade CF, and 1,1,1 TCA resulting in DCM or chloroethane, these end products often persisted and did not undergo further reductive dechlorination. However, a novel enrichment culture derived from pristine river sediment revealed another key dehalogenation pathway associated with Dehalobacter spp. This enrichment culture degraded DCM using fermentative dehalogenation. The resulting end products were acetate, methane, and chloride, which are environmentally benign. | |||
==Isolation and Cultivation== | ==Isolation and Cultivation== | ||
Revision as of 19:15, 18 April 2014
A Microbial Biorealm page on the genus Dehalobacter
Classification
Higher order taxa:
Bacteria; Firmicutes; Clostridia; Clostridiales; Peptococcaceae; Dehalobacter
Species:
Dehalobacter restrictus PER K23, Dehalobacter sp. E1, Dehalobacter sp. TCA1, Dehalobacter sp. MS Dehalobacter sp. WL
|
NCBI: Taxonomy Genome |
Description and Significance
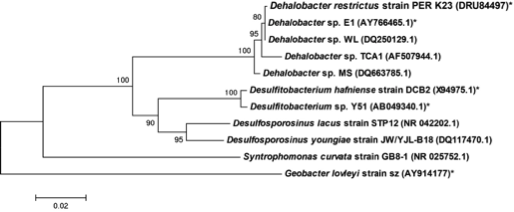
Dehalobacter restrictus is a rod shaped gram-negative obligate anaerobe found in deep soil sediments and groundwater aquifers and is phylogenetically related to several halorespiring genera like Desulfitobacterium and Desulfosporlosinus. It is a typical representative of Dehalobacter spp., which are capable of reductive dechlorination of chlorinated ethenes and chlorinated ethanes (1, 2) and fermentative dehalogenation of Dichloromethane (DCM) (3). These metabolic pathways make them relevant to bioremediation of ground water aquifers contaminated with chlorinated solvents (4). To date, several strains of Dehalobacter have been isolated (1, 3, 5-7)."
Genome Structure
The whole genome of Dehalobacter restrictus PER- K23 was recently sequenced revealing critical insights into phylogeny and metabolic capabilities of the genus. The 2,943 kbp long genome contains 2,826 protein coding and 82 RNA genes, including 5 16S rRNA genes. Interestingly, the genome contains 25 predicted rdase genes, the majority of which appear to be full length. The rdase genes are mainly located in two clusters, suggesting a much larger potential for organohalide respiration than previously anticipated (11).
Cell Structure
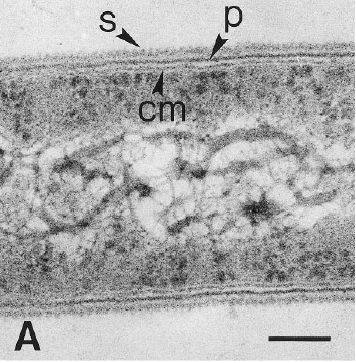
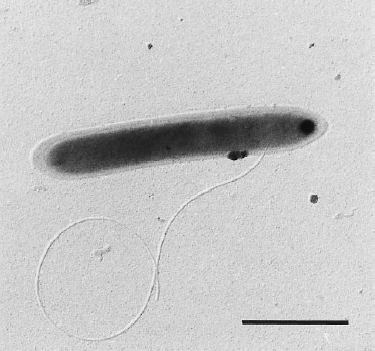
Dehalobacter restrictus cells are rod shaped, 2-3 µm in length and 0.3-0.5 µm in diameter with tapering ends. There is a single lateral flagellum at one end indicating that the cells are motile. The cells do not form spores under substrate and temperature induced stress. Cell wall stains gram negative and is composed of peptidoglycan type A3ϒ enveloped by a layer of proteinaceous surface layer (S – layer) (5)
Metabolism
Dehalobacter restrictus has unique metabolic pathways that make them vital for bioremediation of several key recalcitrant environmental contaminants (4).
Reductive dehalogenation
Reductive dehalogenation is the reduction of halogenated aliphatic and aromatic compounds using hydrogen as an electron donor. Microbially mediated reductive dehalogenation linked to growth is referred to as organohalide respiration. In addition to Dehalobacter restrictus, numerous microbial genera mediate organohalide respiration (e.g. Dehalococcoides, Dehalogimonas, etc.). When a chlorinated molecule is used as electron acceptor for organohalide respiration, and it is thus termed as microbially mediated reductive dechlorination. In this process, the chlorine atom on a chlorinated molecule is replaced by a hydrogen atom using a reductive dehalogenase enzyme (rdase) like pceA.
Dehalobacter restrictus accepts chlorinated solvents, PCE and TCE, as electron acceptors. However, reductive dechlorination is incomplete and stops at cis-1,2 dichlorethene (cDCE). Dehalobacter Strain Known electron acceptors End Products Source PER K23 PCE, TCE cDCE (5) TCA1 1,1,1 TCA, 1,2 DCA Chloroethane (9) MS CF, 1,1,1 TCA, 1,2DCA DCM (10)
Mixed populations of Dehalobacter spp. have also been shown to mediate reductive dehalogenation of 1,2 dichlorobenzene, monochlorobenzene, polychlorinated biphenyl and other recalcitrant chlorinated compounds.
Fermentation of Dichloromethane
Several different strains of Dehalobacter spp. have been known to degrade CF, and 1,1,1 TCA resulting in DCM or chloroethane, these end products often persisted and did not undergo further reductive dechlorination. However, a novel enrichment culture derived from pristine river sediment revealed another key dehalogenation pathway associated with Dehalobacter spp. This enrichment culture degraded DCM using fermentative dehalogenation. The resulting end products were acetate, methane, and chloride, which are environmentally benign.
Isolation and Cultivation
D. restrictus is isolated from a laboratory system that was being fed with tetrachloroethene.