Discovery of Sea Star Associated Densovirus: Difference between revisions
Kspees7838 (talk | contribs) No edit summary |
Kspees7838 (talk | contribs) No edit summary |
||
| Line 25: | Line 25: | ||
Sea stars on the California coast are periodically subject to wasting disease epidemics which leave the populations of many sea star species greatly reduced. Sea stars, particularly serve as keystone predators in their marine ecosystems as they prey on mussels and keep the edges of mussel beds at a remarkably constant location. This work maintains kelp forests which support many other species and aid in protecting the shore. | Sea stars on the California coast are periodically subject to wasting disease epidemics which leave the populations of many sea star species greatly reduced. Sea stars, particularly serve as keystone predators in their marine ecosystems as they prey on mussels and keep the edges of mussel beds at a remarkably constant location. This work maintains kelp forests which support many other species and aid in protecting the shore. | ||
<br>Sea star wasting disease (SSWD) is rather grisly disease which causes sea stars to largely disintegrate and which can occur completely in the course of a few days to a few weeks. The disease begins with a loss of turgor pressure, leading to a deflated appearance, proceeds to the formation of lesions on the sea star, the detachment and “walking away” of individual arms, and, finally, the disintegration and death of the organism. | <br>Sea star wasting disease (SSWD) is rather grisly disease which causes sea stars to largely disintegrate and which can occur completely in the course of a few days to a few weeks. The disease begins with a loss of turgor pressure, leading to a deflated appearance, proceeds to the formation of lesions on the sea star, the detachment and “walking away” of individual arms, and, finally, the disintegration and death of the organism. | ||
<br>As of the summer of 2013, sea stars seem to be undergoing another particularly severe epidemic of this wasting disease. This epidemic is notable for its remarkable spatial spread – from Alaska to Central America, with some accounts from the east coast – and for the diversity of species affected, up to twelve different species currently identified. The ravages of this disease are seen even in remarkably protected environments such as the Monterey Bay Aquarium. The geographic range suggests that there is indeed a pathogenic, instead of environmental, basis to this epidemic. | <br> | ||
<br>As of the summer of 2013, sea stars seem to be undergoing another particularly severe epidemic of this wasting disease. This epidemic is notable for its remarkable spatial spread – from Alaska to Central America, with some accounts from the east coast – and for the diversity of species affected, up to twelve different species currently identified. The ravages of this disease are seen even in remarkably protected environments such as the Monterey Bay Aquarium. The geographic range suggests that there is indeed a pathogenic, instead of environmental, basis to this epidemic. | |||
<br> | |||
<br>In their paper “Sea star disease and population declines at the Channel Islands” Eckert et al outline the extent and background of one such wasting disease epidemic. In a 1997 wasting disease epidemic, incidence of wasting disease at a site ranged from 0.8% to 59.6%” and, a year later, populations of two sea star species had declined 75% and 56%. A few other echinoderm species were also affected. Interestingly, wasting disease epidemics generally seem correlated with marginally warmer water (indeed, affected sea stars removed into cold water recovered in Eckert’s study and the warmer water following El Niño storms seems to have been the ultimate cause, allowing the disease to take its course) which would seem to indicate that this problem may become more frequent and severe as ocean temperatures rise. However, the current wasting disease epidemic confounds researchers due to the current trend of somewhat cooler ocean temperatures on the West Coast. | <br>In their paper “Sea star disease and population declines at the Channel Islands” Eckert et al outline the extent and background of one such wasting disease epidemic. In a 1997 wasting disease epidemic, incidence of wasting disease at a site ranged from 0.8% to 59.6%” and, a year later, populations of two sea star species had declined 75% and 56%. A few other echinoderm species were also affected. Interestingly, wasting disease epidemics generally seem correlated with marginally warmer water (indeed, affected sea stars removed into cold water recovered in Eckert’s study and the warmer water following El Niño storms seems to have been the ultimate cause, allowing the disease to take its course) which would seem to indicate that this problem may become more frequent and severe as ocean temperatures rise. However, the current wasting disease epidemic confounds researchers due to the current trend of somewhat cooler ocean temperatures on the West Coast. | ||
| Line 31: | Line 33: | ||
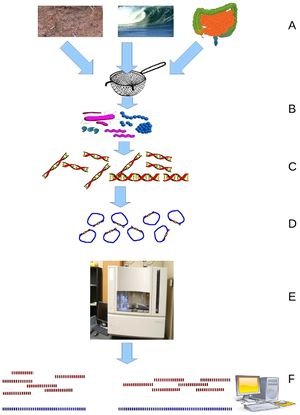
Metagenomics, discussed by Wooley et. al. in his review as the “discipline that enables the genomic study of uncultured microorganisms”, allows researchers to sequence the bulk of bacteria or other microorganisms present in an environmental sample to gain an understanding of what species are present and in what ratios. Considering the impossibility of culturing many microbial species, metagenomics provides a way to gain insight into the diversity of species present. The review also stresses metagenomics’ ability to shed some light on communities of microorganisms and perhaps their interactions with each other or their host which is said to be more relevant to gaining an accurate picture of what is happening than looking at isolated organisms. While powerful, metagenomics is a fairly new and extremely noisy technique, as it relies on researcher’s ability to compile many genomes from many different fragments and researchers cannot expect to find full genomes for the species present. This is made more difficult by the relative newness of whole genome sequencing and the fact that many if not most environmental organisms do not have reference genomes. Metagenomics relies on gene sequencing, most particularly on newer high-throughput sequencing methods such as Roche or Illumina sequencing. The particular challenges of the volume and fragmented nature of the data provided by metagenomics has led to significant work in developing computational strategies to organize the data. | Metagenomics, discussed by Wooley et. al. in his review as the “discipline that enables the genomic study of uncultured microorganisms”, allows researchers to sequence the bulk of bacteria or other microorganisms present in an environmental sample to gain an understanding of what species are present and in what ratios. Considering the impossibility of culturing many microbial species, metagenomics provides a way to gain insight into the diversity of species present. The review also stresses metagenomics’ ability to shed some light on communities of microorganisms and perhaps their interactions with each other or their host which is said to be more relevant to gaining an accurate picture of what is happening than looking at isolated organisms. While powerful, metagenomics is a fairly new and extremely noisy technique, as it relies on researcher’s ability to compile many genomes from many different fragments and researchers cannot expect to find full genomes for the species present. This is made more difficult by the relative newness of whole genome sequencing and the fact that many if not most environmental organisms do not have reference genomes. Metagenomics relies on gene sequencing, most particularly on newer high-throughput sequencing methods such as Roche or Illumina sequencing. The particular challenges of the volume and fragmented nature of the data provided by metagenomics has led to significant work in developing computational strategies to organize the data. | ||
[[File:A-Primer-on-Metagenomics-pcbi.1000667.g001.jpg|thumb|256px wide|Metagenomics Schematic: Environmental samples are filtered down to microbes which from which DNA is isolated and sequenced. From Wooley et. al. A Primer on Metagenomics.]] | [[File:A-Primer-on-Metagenomics-pcbi.1000667.g001.jpg|thumb|256px wide|Metagenomics Schematic: Environmental samples are filtered down to microbes which from which DNA is isolated and sequenced. From Wooley et. al. A Primer on Metagenomics.]] | ||
<br> | |||
<br>There is ample precedent for using metagenomic analyses to gain insight into marine microbial communities. Originally, many different environmental samples were analyzed from the Saragasso Sea which yielded insight into many new marine bacteria and established the effectiveness of using metagenomics approaches to understand marine organisms. Metagenomics has been applied to try to gain insight into a disease, white plague, affecting Mussismilia braziliensis corals. This study could serve almost as a template for approaches to working with metagenomics and marine organisms, instead of general environmental samples. The metagenome was sequenced with Roche sequencing which generated 344,000 sequences with and average length of 414 base pairs. This study saw significant differences in the metagenomes of healthy and sick corals with several different types of bacteria, Bacteroidetes, Cytophaga, Flavobacteria, Rickettsiales, Neisseriales, and Vibrionales, having significantly higher amounts in sick corals and concluded that coral white plague appeared to be a polymicrobial infection. Unfortunately this study did not validate its findings which would have strengthened their postulate. However, this study also took into account other types of organisms, eukaryotes and viruses, which were present in the corals, though they did not focus on them as a source of disease, which would be valuable in expanding the search for the pathogen beyond bacteria. In addition to shedding light on the potential agents behind white plague, this metagenomic analysis provides an initial look into the complexity of the M. braziliensis holobiont through the analysis of levels of known eukaryotic coral symbionts and the relative differences in their amounts between sick and healthy coral. | <br>There is ample precedent for using metagenomic analyses to gain insight into marine microbial communities. Originally, many different environmental samples were analyzed from the Saragasso Sea which yielded insight into many new marine bacteria and established the effectiveness of using metagenomics approaches to understand marine organisms. Metagenomics has been applied to try to gain insight into a disease, white plague, affecting Mussismilia braziliensis corals. This study could serve almost as a template for approaches to working with metagenomics and marine organisms, instead of general environmental samples. The metagenome was sequenced with Roche sequencing which generated 344,000 sequences with and average length of 414 base pairs. This study saw significant differences in the metagenomes of healthy and sick corals with several different types of bacteria, Bacteroidetes, Cytophaga, Flavobacteria, Rickettsiales, Neisseriales, and Vibrionales, having significantly higher amounts in sick corals and concluded that coral white plague appeared to be a polymicrobial infection. Unfortunately this study did not validate its findings which would have strengthened their postulate. However, this study also took into account other types of organisms, eukaryotes and viruses, which were present in the corals, though they did not focus on them as a source of disease, which would be valuable in expanding the search for the pathogen beyond bacteria. In addition to shedding light on the potential agents behind white plague, this metagenomic analysis provides an initial look into the complexity of the M. braziliensis holobiont through the analysis of levels of known eukaryotic coral symbionts and the relative differences in their amounts between sick and healthy coral. | ||
<br> | |||
<br>Most notably, metagenomics has recently been used to identify a densovirus in sea urchins, the first virus discovered to affect echinoderms. Researchers chose to focus on sea urchins because these organisms are important both for aquaculture and maintaining a stable ecosystem and because, while the importance of viruses in marine ecosystems is slowly becoming better known, no viruses affecting echinoderms have previously been characterized. Sea urchin mortality is also known to be affected by other microbial agents and environmental conditions, and a severe epidemic in D. antillarum in the 1980’s followed patterns of a viral outbreak, pointing to the presence of viruses as affecting sea urchin populations. For the metagenomic analysis, samples from three species’ gonads and digestive tract as well as environmental samples were obtained and filtered to remove cells and organisms larger than virus-sized and the absence of bacterial or eukaryotic DNA was confirmed using PCR to amplify 16S or 18S rRNA. The remaining viral DNA was amplified and sequenced with Illumina sequencing and the data was analyzed. Researchers found mainly phages and viruses from the Parvoviridae family and were most interested in the discovery of a densovirus, the form of parvoviridae viruses most likely to infect invertebrates, in sea urchin tissue. Since the urchins investigated did not seem diseased, this seems to be a non-pathogenic virus though evidence of capsid production seemed to confirm that this virus was not endogenous to the host genome. This densovirus was familiar to those affecting arthropods, which allowed researchers to develop probes for the virus and look at its presence in environmental samples which showed this densovirus to be present in sediment and the water column. Metagenomics has thus allowed researchers to identify and characterize novel marine viruses in phyla which previously had no known viruses and attest to the important role which these unknown viruses play in marine populations. | <br>Most notably, metagenomics has recently been used to identify a densovirus in sea urchins, the first virus discovered to affect echinoderms. Researchers chose to focus on sea urchins because these organisms are important both for aquaculture and maintaining a stable ecosystem and because, while the importance of viruses in marine ecosystems is slowly becoming better known, no viruses affecting echinoderms have previously been characterized. Sea urchin mortality is also known to be affected by other microbial agents and environmental conditions, and a severe epidemic in D. antillarum in the 1980’s followed patterns of a viral outbreak, pointing to the presence of viruses as affecting sea urchin populations. For the metagenomic analysis, samples from three species’ gonads and digestive tract as well as environmental samples were obtained and filtered to remove cells and organisms larger than virus-sized and the absence of bacterial or eukaryotic DNA was confirmed using PCR to amplify 16S or 18S rRNA. The remaining viral DNA was amplified and sequenced with Illumina sequencing and the data was analyzed. Researchers found mainly phages and viruses from the Parvoviridae family and were most interested in the discovery of a densovirus, the form of parvoviridae viruses most likely to infect invertebrates, in sea urchin tissue. Since the urchins investigated did not seem diseased, this seems to be a non-pathogenic virus though evidence of capsid production seemed to confirm that this virus was not endogenous to the host genome. This densovirus was familiar to those affecting arthropods, which allowed researchers to develop probes for the virus and look at its presence in environmental samples which showed this densovirus to be present in sediment and the water column. Metagenomics has thus allowed researchers to identify and characterize novel marine viruses in phyla which previously had no known viruses and attest to the important role which these unknown viruses play in marine populations. | ||
==SSaDV== | ==SSaDV== | ||
Sea star wasting disease (SSWD) has been recently analyzed by metagenomics by Hewson’s lab, the same group which investigated viruses in sea urchins, and the results point to a densovirus as the responsible pathogen. Densoviruses belong to the sub-family Densovirinae which, in turn, belongs to the family Parvoviridae. Densovirinae viruses are distinct in that they generally infect invertebrates either in marine or terrestrial ecosystems. This family is undergoing continuous redefinition and division into sub-groups as new and diverse viruses which fit its parameters are characterized. Parvoviridae genomes consist of linear single-stranded DNA and tend to be about 5kb long. They generally end in short palindromic repeats which create hairpin turns which can serve as almost a sort of primer to permit “rolling hairpin” DNA replication. Genomes vary, but universally code for a NS1 gene which initiates replication and serves as a helicase, nickase, and ATPase as well as genes for other structural proteins, VP genes, which form the capsid. | Sea star wasting disease (SSWD) has been recently analyzed by metagenomics by Hewson’s lab, the same group which investigated viruses in sea urchins, and the results point to a densovirus as the responsible pathogen. Densoviruses belong to the sub-family Densovirinae which, in turn, belongs to the family Parvoviridae. Densovirinae viruses are distinct in that they generally infect invertebrates either in marine or terrestrial ecosystems. This family is undergoing continuous redefinition and division into sub-groups as new and diverse viruses which fit its parameters are characterized. Parvoviridae genomes consist of linear single-stranded DNA and tend to be about 5kb long. They generally end in short palindromic repeats which create hairpin turns which can serve as almost a sort of primer to permit “rolling hairpin” DNA replication. Genomes vary, but universally code for a NS1 gene which initiates replication and serves as a helicase, nickase, and ATPase as well as genes for other structural proteins, VP genes, which form the capsid. | ||
<br> | |||
<br>Many researchers expected the pathogen to be a type of vibrio bacteria so these results were surprising. However, the researcher’s initial analysis did not show signs of bacterial infection and virus-sized particles isolated from affected sea stars were sufficient to cause sea star wasting disease in two rounds of previously unaffected individuals, which established the pathogen as a virus. The fact that the disease had spread to sea stars in aquariums indicates that the virus is water-borne and transmission does not rely on contact between affected and unaffected individuals. | <br>Many researchers expected the pathogen to be a type of vibrio bacteria so these results were surprising. However, the researcher’s initial analysis did not show signs of bacterial infection and virus-sized particles isolated from affected sea stars were sufficient to cause sea star wasting disease in two rounds of previously unaffected individuals, which established the pathogen as a virus. The fact that the disease had spread to sea stars in aquariums indicates that the virus is water-borne and transmission does not rely on contact between affected and unaffected individuals. | ||
<br> | |||
<br>The virus was analyzed using metagenomics to compare the viral metagenome of both sick and healthy sea stars. This comparison showed sick sea stars to have a significantly higher level of virus particles from the parvoviridae family. Further analysis of many samples allowed researchers to compile a nearly complete densovirus genome. This virus has been named sea star associated densovirus (SSaDV) and is seemingly the only microbe responsible for sea star wasting disease. This was confirmed by testing multiple species of affected sea stars all of which showed higher levels of SSaDV than any other microbe. qPCR also confirmed that the amounts of virus present in affected sea stars increased throughout the progression of the disease and, across species, individuals with higher levels of SSaDV were more likely to show disease symptoms. Indeed, levels of virus present proved to be the greatest indication for SSWD when compared to factors of location, species, and size. Strikingly, one species, P. ochraceus, showed evidence of the virus both in sick and healthy individuals, indicating a difference in the ways in which different species interact with the virus. Next, environmental samples were investigated to further examine the manner in which SSaDV spreads among individuals. SSaDV was found in particulate matter, indicating that it can travel in the water column to infect new populations, and in other apparently unaffected echinoderm species, indicating a potential reservoir of SSaDV in the ecosystem. Intriguingly, SSaDV has been isolated from diseased sea stars preserved from earlier wasting disease epidemics, which raises questions about why this particular outbreak is so widespread and severe. Researchers speculate that SSaDV could be exacerbated by unidentified environmental factors, overpopulation, or changes in the virus. The definitive discovery of SSaDV is a triumph for metagenomics and an important step in understand SSWD though many questions remain. | <br>The virus was analyzed using metagenomics to compare the viral metagenome of both sick and healthy sea stars. This comparison showed sick sea stars to have a significantly higher level of virus particles from the parvoviridae family. Further analysis of many samples allowed researchers to compile a nearly complete densovirus genome. This virus has been named sea star associated densovirus (SSaDV) and is seemingly the only microbe responsible for sea star wasting disease. This was confirmed by testing multiple species of affected sea stars all of which showed higher levels of SSaDV than any other microbe. qPCR also confirmed that the amounts of virus present in affected sea stars increased throughout the progression of the disease and, across species, individuals with higher levels of SSaDV were more likely to show disease symptoms. Indeed, levels of virus present proved to be the greatest indication for SSWD when compared to factors of location, species, and size. Strikingly, one species, P. ochraceus, showed evidence of the virus both in sick and healthy individuals, indicating a difference in the ways in which different species interact with the virus. Next, environmental samples were investigated to further examine the manner in which SSaDV spreads among individuals. SSaDV was found in particulate matter, indicating that it can travel in the water column to infect new populations, and in other apparently unaffected echinoderm species, indicating a potential reservoir of SSaDV in the ecosystem. Intriguingly, SSaDV has been isolated from diseased sea stars preserved from earlier wasting disease epidemics, which raises questions about why this particular outbreak is so widespread and severe. Researchers speculate that SSaDV could be exacerbated by unidentified environmental factors, overpopulation, or changes in the virus. The definitive discovery of SSaDV is a triumph for metagenomics and an important step in understand SSWD though many questions remain. | ||
Revision as of 01:40, 18 March 2015
Template:BIOL 168L (Microbiology)
From MicrobeWiki, the student-edited microbiology resource
This student page has not been curated.
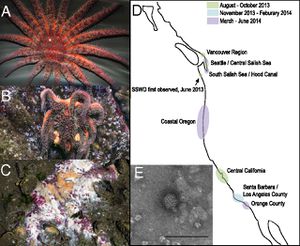
The past year has shown a remarkable epidemic among many sea star species largely centered on the Pacific Coast. This epidemic is notable both because it drastically alters marine ecosystems for which sea stars are keystone predators but also because it is a particularly pervasive and grisly disease – many populations of many species over an impressive geographical distance have been devastated and the disease causes sea stars to eventually just dissolve. New high-throughput sequencing technologies, most notably metagenomics, have begun to yield answers to the source of this epidemic. Researchers were surprised to learn that the infectious agent was a densovirus since it had been widely speculated that the disease was bacterial and caused by vibrio bacteria, well-established marine pathogens. This discovery thus opens up a discussion about newly discovered marine viruses, our expanding ability to detect and characterize them, and their role in marine environments, either natural or used for aquaculture.
Contents [hide] 1 SSWD 2 Metagenomics 3 SSaDV 4 Further Reading 5 References
SSWD
Sea stars on the California coast are periodically subject to wasting disease epidemics which leave the populations of many sea star species greatly reduced. Sea stars, particularly serve as keystone predators in their marine ecosystems as they prey on mussels and keep the edges of mussel beds at a remarkably constant location. This work maintains kelp forests which support many other species and aid in protecting the shore.
Sea star wasting disease (SSWD) is rather grisly disease which causes sea stars to largely disintegrate and which can occur completely in the course of a few days to a few weeks. The disease begins with a loss of turgor pressure, leading to a deflated appearance, proceeds to the formation of lesions on the sea star, the detachment and “walking away” of individual arms, and, finally, the disintegration and death of the organism.
As of the summer of 2013, sea stars seem to be undergoing another particularly severe epidemic of this wasting disease. This epidemic is notable for its remarkable spatial spread – from Alaska to Central America, with some accounts from the east coast – and for the diversity of species affected, up to twelve different species currently identified. The ravages of this disease are seen even in remarkably protected environments such as the Monterey Bay Aquarium. The geographic range suggests that there is indeed a pathogenic, instead of environmental, basis to this epidemic.
In their paper “Sea star disease and population declines at the Channel Islands” Eckert et al outline the extent and background of one such wasting disease epidemic. In a 1997 wasting disease epidemic, incidence of wasting disease at a site ranged from 0.8% to 59.6%” and, a year later, populations of two sea star species had declined 75% and 56%. A few other echinoderm species were also affected. Interestingly, wasting disease epidemics generally seem correlated with marginally warmer water (indeed, affected sea stars removed into cold water recovered in Eckert’s study and the warmer water following El Niño storms seems to have been the ultimate cause, allowing the disease to take its course) which would seem to indicate that this problem may become more frequent and severe as ocean temperatures rise. However, the current wasting disease epidemic confounds researchers due to the current trend of somewhat cooler ocean temperatures on the West Coast.
Metagenomics
Metagenomics, discussed by Wooley et. al. in his review as the “discipline that enables the genomic study of uncultured microorganisms”, allows researchers to sequence the bulk of bacteria or other microorganisms present in an environmental sample to gain an understanding of what species are present and in what ratios. Considering the impossibility of culturing many microbial species, metagenomics provides a way to gain insight into the diversity of species present. The review also stresses metagenomics’ ability to shed some light on communities of microorganisms and perhaps their interactions with each other or their host which is said to be more relevant to gaining an accurate picture of what is happening than looking at isolated organisms. While powerful, metagenomics is a fairly new and extremely noisy technique, as it relies on researcher’s ability to compile many genomes from many different fragments and researchers cannot expect to find full genomes for the species present. This is made more difficult by the relative newness of whole genome sequencing and the fact that many if not most environmental organisms do not have reference genomes. Metagenomics relies on gene sequencing, most particularly on newer high-throughput sequencing methods such as Roche or Illumina sequencing. The particular challenges of the volume and fragmented nature of the data provided by metagenomics has led to significant work in developing computational strategies to organize the data.
There is ample precedent for using metagenomic analyses to gain insight into marine microbial communities. Originally, many different environmental samples were analyzed from the Saragasso Sea which yielded insight into many new marine bacteria and established the effectiveness of using metagenomics approaches to understand marine organisms. Metagenomics has been applied to try to gain insight into a disease, white plague, affecting Mussismilia braziliensis corals. This study could serve almost as a template for approaches to working with metagenomics and marine organisms, instead of general environmental samples. The metagenome was sequenced with Roche sequencing which generated 344,000 sequences with and average length of 414 base pairs. This study saw significant differences in the metagenomes of healthy and sick corals with several different types of bacteria, Bacteroidetes, Cytophaga, Flavobacteria, Rickettsiales, Neisseriales, and Vibrionales, having significantly higher amounts in sick corals and concluded that coral white plague appeared to be a polymicrobial infection. Unfortunately this study did not validate its findings which would have strengthened their postulate. However, this study also took into account other types of organisms, eukaryotes and viruses, which were present in the corals, though they did not focus on them as a source of disease, which would be valuable in expanding the search for the pathogen beyond bacteria. In addition to shedding light on the potential agents behind white plague, this metagenomic analysis provides an initial look into the complexity of the M. braziliensis holobiont through the analysis of levels of known eukaryotic coral symbionts and the relative differences in their amounts between sick and healthy coral.
Most notably, metagenomics has recently been used to identify a densovirus in sea urchins, the first virus discovered to affect echinoderms. Researchers chose to focus on sea urchins because these organisms are important both for aquaculture and maintaining a stable ecosystem and because, while the importance of viruses in marine ecosystems is slowly becoming better known, no viruses affecting echinoderms have previously been characterized. Sea urchin mortality is also known to be affected by other microbial agents and environmental conditions, and a severe epidemic in D. antillarum in the 1980’s followed patterns of a viral outbreak, pointing to the presence of viruses as affecting sea urchin populations. For the metagenomic analysis, samples from three species’ gonads and digestive tract as well as environmental samples were obtained and filtered to remove cells and organisms larger than virus-sized and the absence of bacterial or eukaryotic DNA was confirmed using PCR to amplify 16S or 18S rRNA. The remaining viral DNA was amplified and sequenced with Illumina sequencing and the data was analyzed. Researchers found mainly phages and viruses from the Parvoviridae family and were most interested in the discovery of a densovirus, the form of parvoviridae viruses most likely to infect invertebrates, in sea urchin tissue. Since the urchins investigated did not seem diseased, this seems to be a non-pathogenic virus though evidence of capsid production seemed to confirm that this virus was not endogenous to the host genome. This densovirus was familiar to those affecting arthropods, which allowed researchers to develop probes for the virus and look at its presence in environmental samples which showed this densovirus to be present in sediment and the water column. Metagenomics has thus allowed researchers to identify and characterize novel marine viruses in phyla which previously had no known viruses and attest to the important role which these unknown viruses play in marine populations.
SSaDV
Sea star wasting disease (SSWD) has been recently analyzed by metagenomics by Hewson’s lab, the same group which investigated viruses in sea urchins, and the results point to a densovirus as the responsible pathogen. Densoviruses belong to the sub-family Densovirinae which, in turn, belongs to the family Parvoviridae. Densovirinae viruses are distinct in that they generally infect invertebrates either in marine or terrestrial ecosystems. This family is undergoing continuous redefinition and division into sub-groups as new and diverse viruses which fit its parameters are characterized. Parvoviridae genomes consist of linear single-stranded DNA and tend to be about 5kb long. They generally end in short palindromic repeats which create hairpin turns which can serve as almost a sort of primer to permit “rolling hairpin” DNA replication. Genomes vary, but universally code for a NS1 gene which initiates replication and serves as a helicase, nickase, and ATPase as well as genes for other structural proteins, VP genes, which form the capsid.
Many researchers expected the pathogen to be a type of vibrio bacteria so these results were surprising. However, the researcher’s initial analysis did not show signs of bacterial infection and virus-sized particles isolated from affected sea stars were sufficient to cause sea star wasting disease in two rounds of previously unaffected individuals, which established the pathogen as a virus. The fact that the disease had spread to sea stars in aquariums indicates that the virus is water-borne and transmission does not rely on contact between affected and unaffected individuals.
The virus was analyzed using metagenomics to compare the viral metagenome of both sick and healthy sea stars. This comparison showed sick sea stars to have a significantly higher level of virus particles from the parvoviridae family. Further analysis of many samples allowed researchers to compile a nearly complete densovirus genome. This virus has been named sea star associated densovirus (SSaDV) and is seemingly the only microbe responsible for sea star wasting disease. This was confirmed by testing multiple species of affected sea stars all of which showed higher levels of SSaDV than any other microbe. qPCR also confirmed that the amounts of virus present in affected sea stars increased throughout the progression of the disease and, across species, individuals with higher levels of SSaDV were more likely to show disease symptoms. Indeed, levels of virus present proved to be the greatest indication for SSWD when compared to factors of location, species, and size. Strikingly, one species, P. ochraceus, showed evidence of the virus both in sick and healthy individuals, indicating a difference in the ways in which different species interact with the virus. Next, environmental samples were investigated to further examine the manner in which SSaDV spreads among individuals. SSaDV was found in particulate matter, indicating that it can travel in the water column to infect new populations, and in other apparently unaffected echinoderm species, indicating a potential reservoir of SSaDV in the ecosystem. Intriguingly, SSaDV has been isolated from diseased sea stars preserved from earlier wasting disease epidemics, which raises questions about why this particular outbreak is so widespread and severe. Researchers speculate that SSaDV could be exacerbated by unidentified environmental factors, overpopulation, or changes in the virus. The definitive discovery of SSaDV is a triumph for metagenomics and an important step in understand SSWD though many questions remain.
Further Reading
[Sample link] Ebola Hemorrhagic Fever—Centers for Disease Control and Prevention, Special Pathogens Branch
References
[Sample reference] Takai, K., Sugai, A., Itoh, T., and Horikoshi, K. "Palaeococcus ferrophilus gen. nov., sp. nov., a barophilic, hyperthermophilic archaeon from a deep-sea hydrothermal vent chimney". International Journal of Systematic and Evolutionary Microbiology. 2000. Volume 50. p. 489-500.
Edited by Kaitlyn Spees, a student of Nora Sullivan in BIOL168L (Microbiology) in The Keck Science Department of the Claremont Colleges Spring 2014.
Categories: ##Uncurated Pages
- Pages edited by students of Nora Sullivan at the Claremont Colleges