User:S4353567: Difference between revisions
No edit summary |
No edit summary |
||
| Line 5: | Line 5: | ||
[[File:Porphyromonas_gingivalis.png|frame|SEM (scanning electron microscopy) image of Porphyromonas ''gingivalis'' <sup>[[#References|[19]]]</sup>]] | [[File:Porphyromonas_gingivalis.png|frame|SEM (scanning electron microscopy) image of Porphyromonas ''gingivalis'' <sup>[[#References|[19]]]</sup>]] | ||
<font size=" | <font size="5">'''Porphyromonas ''gingivalis'''''</font> | ||
==Classification== | ==Classification== | ||
Latest revision as of 13:21, 23 September 2016
Kiranjot Kaur Bench C 43535675 [1]
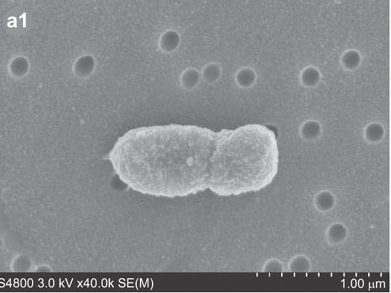
Porphyromonas gingivalis
Classification
Higher order taxa
Eubacteria (Kingdom) – Bacteria (Domain) – Bacteroidetes (Phylum) – Bacteroidia (Class) – Bacteroidales (Order) – Prophyeomonadaceae (Family) – Prophyromonas (Genus)[1]
Species
Porphyromonas gingivalis Type Strain ATCC 33277 [2]
Description and significance
Porphyromonas gingivalis, previously named Bacteriodes gongivalis, is a gram negative bacterium which occurs as coccobacilli or as long rods (0.5 by 1.0 to 2.0 microm).[3] This organism is a major pathogen in chronic periodontitis.[4] Although it is a natural member of the human oral microbiome it can become highly destructive and proliferate rapidly and is able to destroy periodontal tissues causing disease.[3, 4] However, P. gingivalis is detected in very low numbers in healthy individuals.[4] It is a secondary colonizer of dental plaque.[4]
It was first isolated by Slots from a human gingival sulcus. P. gingivalis has an absolute requirement for iron for its growth and is able to be cultured on blood agar with hemin and menadione.[3] It forms small 1 to 2mm convex colonies with brown to black pigment thought be the result of the accumulation of hemin.[3,5,7] P. gingivalis is a fermentative organism.[3,5]
Genome structure
Using the type strain P. gingivalis has a circular genome with no plasmids found.[7] It contains 2,354,866 nucleotides with 2154 genes and of those 2089 are protein encoding genes and 65 RNA encoding genes.[7] This strain contains no pseudogenes and the DNA base composition is 46.5 to 48.4 mol% of G+C.[3] The 16S rRNA sequence for this strain is X73964.[7] P. gingivalis human isolates are both phenotypically and genetically distinct from those recovered from other mammals. [8]
Cell structure and metabolism
P. gingivalis, being a gram negative bacterium it contains a cell wall with peptidoglycan with lysine as the diamino acid.[3,4] This bacteria contains fimbriae involved in cell invasion along with hemaglutinins and proterinases.[7]
P. gingivalis is a non-motile organism, obligate anaerobe and utilises iron and protoporphyrin IX for its growth.[9] Protoporphyrin IX is a carrier for divalent cations and is an essential prosthetic group for heme.[9] It can be cultured in the presence of heme and its growth is markedly affected by the presence of protein hydrolysates.[3,7] P. gingivalis contains five major hemagglutinin genes (hagA, hagB, hagC, hagD and hagE) allowing for absorption, invasion of the bacteria in to cells as well as iron utilisation.[7]
P. gingivalis utilises several amino acids for its growth such as aspartate, arginine, cystine, histidine, serine, tryptophan, etc.[3] It has fermentation products such as n-butyric, isobutyric, propionic, iso-valeric and acetic acids.[3] They are also able to produce low levels of other acids such as phenylacetic acids. Proteases like collagenase are detected in its metabolites.[3]
P. gingivalis forms complex biofilms.[4,7,10]It is unable to colonise clean tooth surfaces. However it will colonise inflammation site as well as sites that contain gram-positive dental plaque bacteria such as Streptococcus gordonii, which is a primary coloniser on teeth.[4,7,9] This provides an attachment site as well as supplying growth substrates.[4,7,10]
Ecology
P. gingivalis is an obligate anaerobic bacterium that resides in the oral cavity. It is a natural member of the oral microbiome and is a late coloniser.[9] It is found in close proximity to and interacts with juxtaposing gingival tissue.[4] It is established in the periodontal pocket. According to the World health organisation periodontal disease affects 10 to 15% of adults worldwide.[4]
P. gingivalis is able to invade cells, such as epithelial cells, and tissues which allows them to replicate and survive.[4] Invasion is allowed to happen with the aid of major fimbriae which binds to the β1 integrin on the host cell which causes the rearrangement of the actin cytoskeleton to allow internalisations. [4] However when they do invade they do know that they do not initiate apoptosis or necrosis. [4] To allow survival P. gingivalis secrete ATP-hydrolysing enzyme which suppresses ATP-dependent apoptosis. [4] It is also able to spread through cells via actin cytoskeleton bridges. [4] This allows spread without causing host cell death as well as avoiding immune surveillance. Once inside P. gingivalis affects cell-cycle pathways accelerating proliferation of gingival epithelia cells. [4]
Pathology
P. gingivalis is a major pathogen in periodontitis and is part of sublingual plaque. The strain differences in P. gingivalis can influence virulence.[8] This pathogen has also been linked in with systemic diseases such as cardiovascular diseases. [4,6,7,8] It is able to invade epithelial, endothelial and smooth muscular cells. [4,8]
Virulence factors allow this pathogen to become opportunistic.[4,6,7,8] The virulence factors elicit deleterious effects on the host.[4,6] Virulence factors that are present within P. gingivalis are lipopolysaccharide (LPS), capsular polysaccharide (CPS), fimbriae and gingipains.[4,7,10]
LPS is recognised by the Toll like receptors (TLRs), is a stimulator of proinflammatory responses and it also stimulates inflammatory cytokine production such as interleukins and tumour necrosis factors (TNF).[4] Lipid A of P. gingivalis activates TLR2 which dampens the immune response allowing for survival.[4] The CPS or K-antigen is considered to be the capsule of the P. gingivalis and it is able to generate IgG antibody responses.[4] The encapsulated P. gingivalis stains are highly invasive which cause the spread of infection.[4] The fimbriae are filamentious cell surface protrusions that allow for adherence to different types of structures on host cells. Fimbriae also allows for attachment to early colonising bacteria a facilitate in the development of biofilm structure.[4] Gingipains has multiple effects such as deregulating immune responses by stimulting IL-6 etc., confering resistance to P. gingivalis bacterial activity.[4] They also affect vascular permeability and bleeding at the periodontal site.[4] P. gingivalis also inhibits IL-8 accumulation and this had drastic innate immune defense in the periodontium.[4] This results in the decreased capacity of directing leukocytes for the removal of bacteria that are present at these sites, resulting in bacterial overgrowth.[4]
It can be aided in entering in to the circulation of the body via eating. P. gingivalis is able to invade cardiovascular cells invasion of cells allows access to host proteins and iron, which are essential for its survival.[8] It has been found to be present in artheromas of the circulatory system. Invasion allows persistence of the pathogen in infected tissue allowing to avoid humoral and cellular responses.[8] Intracellular survival also provides protection against antibiotics.[8]
An enzyme expressed by P. gingivalis, peptidylarginine deiminase (PAD), had been associated with increased risk of developing rheumatoid arthritis (RA).[12]
Application to biotechnology
P. gingivalis has been used in understanding biofilm formation and how this bacterium interacts with other bacterium to understand which proteins on its cells can be altered as well as transport proteins i.e increased or decreased. Allowing for understanding of biofilm formation.[13]
Virulence factors of P. gingivalis are important in developing treatment of periodontal disease. Such as proteases that hydrolyse tissue proteins and eventually leading to tissue destruction. Thus these enzymes are possible targets for drug treatments. Since this pathogen is able to evade the immune system quite effectively through its molecules, these virulence factors are of great interest since they all can be potential drug targets, for example DX-9065a is able to inhibit P. gingivalis growth because it is able to inhibit trypsin-like proteinase activity.[14] This could potentially be a possible target for treatment for adult periodontitis.
This organism is also of interest since it has been linked to other diseases such as rheumatoid arthritis and cardiovascular diseases and so its interaction with host immune cells and its ability to mimicry leading to pathogenesis in the host is another application.[10,13] Understanding this will allow for potential drug targets to be identified and possibly develop drugs to prevent and treat diseases caused by P. gingivalis.
Current research
In current research there has been increased in interest in plant-derived natural products in order to understand their therapeutic roles in regulating the interactions between microorganisms.[15] These compounds seem to be non toxic. There is also research to suggest silicone nitride may be a beneficial treatment of acute or chronic periodontitis. Current research has also found that P. gingivalis may be responsible for rheumatoid arthritis. P. gingivalis eptidyl-arginine deiminase (PPAD) has been found to citrullinates host-derived or bacterial proteins in the inflammatory sites of periodontitis.[10,12] Susceptible individuals start to develop antibodies which bind to the citrullinated proteins resulting in an inflammatory immune response causing PA. This is an indication of autoimmunity against citrullinated proteins. The outer membrane vesicles of P. gingivalis may also contribute to the tissue destruction which contain antigens and proteases. HmuY, another virulence factor of P. gingivalis may be important in growth in heme-limited host environment and infection of macrophages.[16]
It is been found that at least 95% of clinical stains of P. gingivalis carry CRISPR array and this is important in this organism since it is active and possibly provides protection from foreign genetic elements.[17] This research can further the understanding of bacterial community interactions and possibly the process in which disease develops and progresses.
Current research is also looking in to the symbiosis of the different bacterial species within the human oral microbiome. Further studies are looking in to prevention of biofilm formation on teeth.[13]
References
2. LPSN: List of Prokaryotic names with Standing in Nomenclature
3. Shah H.N, Collins M.D. (1988) Proposal for Reclassification of Bacteroides asaccharolyticus, Bacteroides gingivalis, and Bacteroides endodontalis in a New Genus, Porphyromonas. Int J Syst Bacteriol 38:128-131.
4. Bostanci N., Belibasakis G.N. (2012) Porphyromonas gingivalis: an invasive and evasive opportunistic oral pathogen. FEMS microbiology letters 333:1-9.
5. Coykendall A.L., Kaczmarek F.S., and Slots J. (1980) Genetic heterogeneity in Bacteroides asaccharolyticus (Holdeman and Moore 1970) Finegold and Barnes 1977 (Approved Lists, 1980) and proposal of Bacteroides gingivalis sp. nov. and Bacteroides macacae (Slots and Genco) comb. nov. Int J Syst Bacteriol 30:559-564.
6. Summary of Porphyromonas gingivalis, Strain ATCC 33277
7. How K, Song K, Chan K. (2016) Porphyromonas gingivalis: An Overview of Periodontopathic Pathogen below the Gum Line, vol 7, p 1-14.
8. Olsen I., and Progulske-Fox A., (2015) Invasion of Porphyromonas gingivalis strains into vascular cells and tissue. Journal of Oral Microbiology 7:10.3402/jom.v3407.28788.
9. Olczak T, Simpson W, Liu X, Genco CA. (2005) Iron and heme utilization in Porphyromonas gingivalis, vol 29, p 119-144, Oxford, UK.
10. Hajishengallis G. 2015. Periodontitis: from microbial immune subversion to systemic inflammation. Nat Rev Immunol 15:30-44.
11. Darveau RP, Belton CM, Reife RA, Lamont RJ. 1998. Local Chemokine Paralysis, a Novel Pathogenic Mechanism for Porphyromonas gingivalis. Infect Immun 66:1660-1665.
12. Maresz KJ, Hellvard A, Sroka A, Adamowicz K, Bielecka E, Koziel J, Gawron K, Mizgalska D, Marcinska KA, Benedyk M, Pyrc K, Quirke A-M, Jonsson R, Alzabin S, Venables PJ, Nguyen K-A, Mydel P, Potempa J. (2013) Porphyromonas gingivalis Facilitates the Development and Progression of Destructive Arthritis through Its Unique Bacterial Peptidylarginine Deiminase (PAD). PLoS Pathog 9:e1003627.
13. Mysak J, Podzimek S, Sommerova P, Lyuya-Mi Y, Bartova J, Janatova T, Prochazkova J, Duskova J. (2014) Porphyromonas gingivalis: Major Periodontopathic Pathogen Overview. Journal of Immunology Research 2014:8.
14. Matsushita K, Imamura T, Tancharoen S, Tatsuyama S, Tomikawa M, Travis J, Potempa J, Torii M and Maruyama I. (2006) Selective inhibition of Porphyromonas gingivalis growth by a factor Xa inhibitor, DX-9065a. Journal of Periodontal Research 41:171-176.
15. Grenier D. and Dang La V. (2011) Proteases of Porphyromonas gingivalis as Important Virulence Factors in Periodontal Disease and Potential Targets for Plant-Derived Compounds: A Review Article. Current Drug Targets 12:322-331.
16. Carvalho-Filho P.C., Gomes-Filho I.S., Meyer R., Olczak T., Xavier M.T., Trindade S.C. (2016) Role of Porphyromonas gingivalis HmuY in Immunopathogenesis of Chronic Periodontitis. Mediators of Inflammation 2016:7465852.
17. Burmistrz M, Dudek B, Staniec D, Rodriguez Martinez JI, Bochtler M, Potempa J, Pyrc K. 2015. Functional Analysis of Porphyromonas gingivalis W83 CRISPR-Cas Systems. J Bacteriol 197:2631-2641.
18. Ang Cs, Veith PD, Dashper SG, Reynolds EC. (2008) Application of 16 O/ 18 O reverse proteolytic labeling to determine the effect of biofilm culture on the cell envelope proteome of Porphyromonas gingivali s W50. Proteomics 8:1645-1660.
19. Fischer CL, Walters KS, Drake DR, Dawson DV, Blanchette DR, Brogden KA, Wertz PW. (2013) Oral mucosal lipids are antibacterial against Porphyromonas gingivalis, induce ultrastructural damage, and alter bacterial lipid and protein compositions. International Journal of Oral Science 5:130-140.
- ↑ MICR3004
This page is written by Kiranjot Kaur for the MICR3004 course, Semester 2, 2016
