Moritella marina: Difference between revisions
| Line 26: | Line 26: | ||
[[image:16s.png|thumbnail|300px| Figure 2: Image credit:(Urakawa | [[image:16s.png|thumbnail|300px| Figure 2: Image credit:(Urakawa 2014)]] | ||
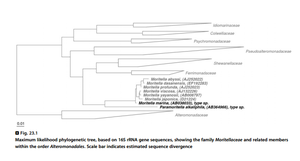
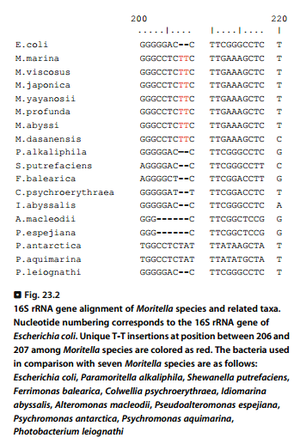
Based on 16S rRNA gene analysis, ''Moritella marina'' shows nearly identical similarities with other ''Moritella'' species (97-99% identity with other species). Unique T-T insertions at position between 206 and 207 among ''Moritella'' species can help to differentiate ''Moritella'' species from other closely related taxa (Fig. 2). | Based on 16S rRNA gene analysis, ''Moritella marina'' shows nearly identical similarities with other ''Moritella'' species (97-99% identity with other species). Unique T-T insertions at position between 206 and 207 among ''Moritella'' species can help to differentiate ''Moritella'' species from other closely related taxa (Fig. 2). | ||
Revision as of 04:07, 14 December 2016
Classification
Domain: Bacteria
Phylum: Proteobacteria
Class: Gammaproteobacteria
Order: Alteromonadales
Family: Moritellaceae
Genus: Moritella
Species
Moritella marina ATCC 15381T
Related species M. japonica, M. yayanosii, M. viscosa, M. profunda, M. abyssi, M. dasanesis
Description and Significance
Moritella marina is a gram-negative halophilic psychrophilic facultative anaerobe with curved or straight rods, motile polar flagella that produce PUFA’s (polyunsaturated fatty acids) and DHA (docosahexanoic acid). “Twenty-five years ago, a marine bacterium originally designated Vibrio marinus and later renamed Moritella marina MP-1 was reported to produce high levels of DHA (18% of the total fatty acids). Such high levels of DHA presumably provide this bacterium with the ability to maintain appropriate membrane fluidity in the low temperatures and high pressures of its marine environment.”
16S Ribosomal RNA Gene Information
Based on 16S rRNA gene analysis, Moritella marina shows nearly identical similarities with other Moritella species (97-99% identity with other species). Unique T-T insertions at position between 206 and 207 among Moritella species can help to differentiate Moritella species from other closely related taxa (Fig. 2).
Genome Structure
The draft genome of M. marina MP-1 (ATCC 15381) consisting of 83 contigs includes 4,636,778 bp and 4,121 predicted coding DNA sequences with a G+C content of 40.5%. There are 34 predicted RNAs, one predicted copy of 16S rRNA, and four predicted copies of 23S rRNA (Kautharapu and Jarboe 2012).
Ecology and pathogenesis
Moritella marina has been found in a variety of cold marine environments, ranging from the ocean floor to the intestinal tract of marine organisms. Most Moritella species are thought to live mutualistically amongst marine organisms, but Moritella viscosa has been noted to cause skin ulcers in some cold water fish (Urakawa 2014).
Current Research
Moritella marina has been noted for having unusually high production of the long chain polyunsaturated fatty acids (PUFAs). When M. marina strain MP-1 was cultured in medium containing cerulenin, a fatty acid synthesis inhibitor, decreases in levels of middle-chain fatty acids and remarkable increases in levels of DHA were observed. These results suggest that the synthesis of middle-chain fatty acids works independently of the synthesis of DHA. M. marina was also found to produce chitonase when induced with chiton (Stefanidi and Vorgias 2008).
References
Kautharapu, K. B., & Jarboe, L. R. (2012). Genome sequence of the psychrophilic deep-sea bacterium Moritella marina MP-1 (ATCC 15381). Journal of Bacteriology, 194(22), 6296-6297. http://jb.asm.org/content/194/22/6296.full
Pukall, R., Päuker, O., Buntefuß, D., Ulrichs, G., Lebaron, P., Bernard, L., ... & Stackebrandt, E. (1999). High sequence diversity of Alteromonas macleodii-related cloned and cellular 16S rDNAs from a Mediterranean seawater mesocosm experiment. FEMS Microbiology Ecology, 28(4), 335-344.
http://femsec.oxfordjournals.org/content/28/4/335
Stefanidi, E., & Vorgias, C. E. (2008). Molecular analysis of the gene encoding a new chitinase from the marine psychrophilic bacterium Moritella marina and biochemical characterization of the recombinant enzyme. Extremophiles, 12(4), 541-552.
Urakawa H. (2014). The family Moritellaceae. In The Prokaryotes, 4th edition. Edited by Eugene Rosenberg, Edward F. DeLong, Fabiano Thompson, Stephen Lory and Erko Stackebrandt. Springer. New York. Gammaproteobacteria pp. 477-489.
Urakawa, H., Kita-Tsukamoto, K., Steven, S. E., Ohwada, K., & Colwell, R. R. (1998). A proposal to transfer Vibrio marinus (Russell 1891) to a new genus Moritella gen. nov. as Moritella marina comb. nov. FEMS Microbiology Letters, 165(2), 373-378. http://femsle.oxfordjournals.org/content/165/2/373.full#ref-13
Author
Sarah Fayed and Brandon Galindo with Microbial Ecology Instructor Dr. Hideotoshi Urakawa