Streptomyces coelicolor: Difference between revisions
| Line 41: | Line 41: | ||
==Description and significance== | ==Description and significance== | ||
''Streptomyces coelicolor'', a filamentous, high G-C, gram + bacteria, was first dubbed ''Streptothrix coelicolor'' in 1908 by R. Muller after he found it on a potato(2). Later, it became known as ''Streptomyces coelicolor''. The ''Streptomyces coelicolor'' | ''Streptomyces coelicolor'', a filamentous, high G-C, gram + bacteria, was first dubbed ''Streptothrix coelicolor'' in 1908 by R. Muller after he found it on a potato(2). Later, it became known as ''Streptomyces coelicolor''. The ''Streptomyces coelicolor'' | ||
A3(2) strain studied in depth by David A Hopwood and sequenced by the John Innes Center and the Sanger Institute is actually taxonomically a member of the ''Streptomyces violaceoruber | A3(2) strain studied in depth by David A Hopwood and sequenced by the John Innes Center and the Sanger Institute is actually taxonomically a member of the ''Streptomyces violaceoruber'' genus, although it retains the former name, and is not the same strain as the Muller strain(25). ''Streptomyces coelicolor'', like the streptomyces genus in general, live in the soil. Streptomyces are responsible for much of the break down of organic material in the soil as well as the “earthy” smell of soil. They also live in colonies and have structural similarities to fungus. Colonies of ''Streptomyces coelicolor'' release pigments that are blue/green in alkali and red in acidic conditions, thereby giving the bacterial colonies those colors under the respective conditions. Other differentiating characteristics of Muller's ''Streptomyces coelicolor'' are grayish-yellow aerial mycelium, smooth spores, and no melanoid pigment(5). ''Streptomyces coelicolor'' are important bacteria and were sequenced because of their “adaptability to environmental stress”, “source of bioactive molecules for medicine and industry”, and “relat[ion] to human pathogens”(3). ''Streptomyces coelicolor'' has a very similar core genome to ''Mycobacterium tuberculosis'' and ''Corynebacterium diphtheriae'', as well as some similarity to ''Mycobacterium leprae'', so it can be used to study these disease causing bacteria(4). The streptomyces genus is responsible for producing a majority of the antibiotics in use today, as well as some immunosuppressants and anti-tumor agents. ''Streptomyces coelicolor'' also has an interesting life-cycle that includes differentiation into aerial mycelium and spore formation(3). | ||
'' genus, although it retains the former name, and is not the same strain as the Muller strain. ''Streptomyces coelicolor'', like the streptomyces genus in general, live in the soil. Streptomyces are responsible for much of the break down of organic material in the soil as well as the “earthy” smell of soil. They also live in colonies and have structural similarities to fungus. Colonies of ''Streptomyces coelicolor'' release pigments that are blue/green in alkali and red in acidic conditions, thereby giving the bacterial colonies those colors under the respective | |||
Edited by Amy Stapp, student of Rachel Larsen at UCSD. | Edited by Amy Stapp, student of Rachel Larsen at UCSD. | ||
Revision as of 00:14, 5 June 2007
A Microbial Biorealm page on the genus Streptomyces coelicolor
Classification
===Higher order taxa===
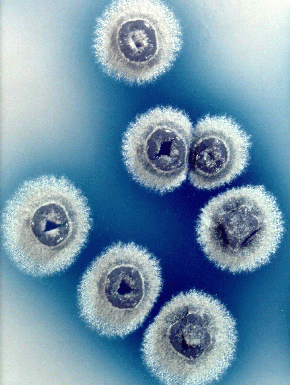
Domain: Bacteria
Phylum: Actinobacteria
Class: Actinobacteria
Subclass: Actinobacteridae
Order: Actinomycetales
Suborder: Streptomycineae
Family: Streptomycetaceae
Strains: Streptomyces coelicolor A3(2)
|
NCBI: Taxonomy |
(1)
Genus
Genus species: Streptomyces coelicolor
Other Names: Streptothrix coelicolor, Cladothrix coelicolor, Nocardia coelicolor, Actinomyces coelicolor
|
NCBI:Taxonomy |
(1)
Description and significance
Streptomyces coelicolor, a filamentous, high G-C, gram + bacteria, was first dubbed Streptothrix coelicolor in 1908 by R. Muller after he found it on a potato(2). Later, it became known as Streptomyces coelicolor. The Streptomyces coelicolor A3(2) strain studied in depth by David A Hopwood and sequenced by the John Innes Center and the Sanger Institute is actually taxonomically a member of the Streptomyces violaceoruber genus, although it retains the former name, and is not the same strain as the Muller strain(25). Streptomyces coelicolor, like the streptomyces genus in general, live in the soil. Streptomyces are responsible for much of the break down of organic material in the soil as well as the “earthy” smell of soil. They also live in colonies and have structural similarities to fungus. Colonies of Streptomyces coelicolor release pigments that are blue/green in alkali and red in acidic conditions, thereby giving the bacterial colonies those colors under the respective conditions. Other differentiating characteristics of Muller's Streptomyces coelicolor are grayish-yellow aerial mycelium, smooth spores, and no melanoid pigment(5). Streptomyces coelicolor are important bacteria and were sequenced because of their “adaptability to environmental stress”, “source of bioactive molecules for medicine and industry”, and “relat[ion] to human pathogens”(3). Streptomyces coelicolor has a very similar core genome to Mycobacterium tuberculosis and Corynebacterium diphtheriae, as well as some similarity to Mycobacterium leprae, so it can be used to study these disease causing bacteria(4). The streptomyces genus is responsible for producing a majority of the antibiotics in use today, as well as some immunosuppressants and anti-tumor agents. Streptomyces coelicolor also has an interesting life-cycle that includes differentiation into aerial mycelium and spore formation(3).
Edited by Amy Stapp, student of Rachel Larsen at UCSD.
Genome structure
Streptomyces coelicolor has one linear chromosome and two plasmids, one that is linear and one that is circular. The linear chromosome was sequenced from overlapping clones of the species, most of which were cosmids, that did not contain the two plasmids. This chromosome contains 8,667,507 base pairs, and was the largest bacterial genome sequenced at the time. (Since then, larger bacterial genomes have been sequenced.) The origin of replication (oriC) is located in the middle of the chromosome, and the ends of the chromosome contain terminal inverted repeats (TIRs). The 5’ terminal ends have proteins that are covalently bonded to them. Replication occurs in both directions leaving a gap in one strand of the new chromosome, which is patched by DNA synthesis. The chromosome is considered to be grouped into three regions – the core and two arms. The core region comprises about half of the chromosome and contains the essential genes for the survival of the organism, like “cell division, DNA replication, transcription, translation and amino-acid biosynthesis” (6). The two arm regions are different lengths, one about 1.5 MB and the other 2.3 MB long, and they code for nonessential functions like "secondary metabolites, hydrolytic exoenzymes, the conservons (conserved operons) and 'gas vesicle' proteins" (6). The SPC1 linear plasmid is 365,023 base pairs long, and is involved coding for some regulator proteins including three Sigma factors and proteins found on spore surfaces among other functions(7). The 31,317 base pair, circular plasmid, SPC2, has a stability region, replication origin, and transfer region. It has a relatively low copy number(8).
Edited by Amy Stapp, student of Rachel Larsen at UCSD.
Cell structure and metabolism
Streptomyces coelicolor live in the soil, where nutrient conditions can change dramatically. As a result, this bacteria is capable of living on may different carbon sources including "glycerol, L-arabinose, D-arabitol, D-ribose, D-xylose, L-xylitol, D-fructose, D-galactose, D-gluconate, D-glucose, D-mannitol, D-mannose, L-rhamnose, salicin, cellobiose, lactose, maltose, melibiose, trehalose, acetate, citrate, lactate, malate, pyruvate, succinate, tartrate, propanoate, alanine, asparagine, glutamate, glycine, histidine, isoleucine, leucine, lysine, methionine, proline, serine, threonine, tryptophan, [and] valine"(11). Some of its nitrogen sources are "aspartate, asparagine, glutamate, glutamine, isoleucine, leucine, lysine, praline, [and] valine"(11). Despite Streptomyces coelicolor ability to metabolize so many different food sources, it is an obligate aerobe. The lactate dehydrogenase gene is present in Streptomyces coelicolor genome, so the organism should be able to obtain energy from fermentation, but it does not. Some theories as to why this occurs are that some of the other reactions necessary for survival depend on the presence of oxygen or that the byproducts of anaerobic respiration are toxic to the cells(11). The presence of nar genes, which code for respiratory nitrate reductaces, indicate that under oxygen limiting conditions, Streptomyces coelicolor should be able to use nitrate as an electron receptor. Researchers have determined that the nar genes are indeed expressed and probably used during growth in standing liquid where oxygen levels fluctuate. Despite these findings, conditions that would allow Streptomyces coelicolor to grow anaerobically in a lab setting have not been found yet(16). Metabolic changes in Streptomyces coelicolor also affect cell differentiation. For example, mutant strains lacking citA, involved in citrate synthase coding, or some bld genes (discussed below) cannot form aerial mycelium when grown on glucose. Glucose forms acidogenic organic acids which makes the substrate in which the organism grows acidic. Citrate synthase initiates the TCA cycle which is necessary for acid metabolism, which is important in keeping the pH of the substrate at a level that does not prevent growth. Without the citA gene or some of the bld genes, this important ability in impaired and, as such, aerial mycelium and antibiotic production do not occur. When grown on mannitol, which is not acidogenic, aerial hyphae will form even in bld and citA mutants(15).
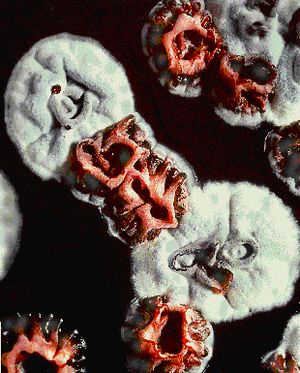
Streptomyces have a life cycle similar to that of fungi. The cycle starts with growth of vegetative mycelium from a spore, followed by ariel mycelium, and, then, spores. This complex life style is facilitated by cell differentiation. Instead of two totally separated cells forming after cell division, chains of cells remain linked together to form a branching mycelium network. Vegetative hyphae, the individual strands that make up the vegetative mycelium, have hydrophilic surfaces, which fit well since they usually grown in a moist region. When not grown in submerged culture, hyphae have an extra cellular matrix of up to 1.5 micrometers wide that surround it(13). Older hyphae have larger extra cellular layers than newer hyphae. Production of aerial mycelium is coded for in the bld genes of Streptomyces coelicolor. When grown in the presence of glucose, SapB is believed to help aerial mycelium break the surface tension of the liquid they begin growing in and ascend into the air(12). The surfaces of aerial hyphae, the individual strands that make up the aerial mycelium, are hydrophobic, unlike vegetative hyphae. A fibrous layer also surrounds newly formed aerial hyphae, and is believed to help break surface tension as the hyphae move out of an aqueous substrate and into the air. This fibrous layer is not present in older aerial hyphae, so it is believed to be replaced with a “more organized mosaic layer”(13). Sporulation in Streptomyces coelicolor is controlled by the whi genes. Sporulation begins when indentations at the tips of aerial hyphae begin to appear. These indentations are the place where spores will form. The mosaic of fibers covering the aerial hyphae then form a ring around the indentations. A round spore then forms and is covered in the fibrous mosaic. Mature spores are denoted by surface concavities believed to be caused by metabolic slowing and dehydration(13).
Edited by Amy Stapp, student of Rachel Larsen at UCSD.
Ecology
Streptomyces coelicolor and other Streptomyces species are important to soil environments because they are capapble of metabolizing other organism's remains. They are espescially important because they can degrade chitin and other compounds that are difficult to degrade(19). This ability makes them an integral part of the global carbon cycle.
Streptomyces coelicolor also takes part in the nitrogen cycle. Several nar genes, as well as a few others, code for the products necessary to reduce nitrate to nitrite. Nitrite is reduced to ammonia by products coded for in nir genes as well. The role of decomposers, like Streptomyces coelicolor, as nitrogen reducers is a major step in the nitrogen cycle.(24)
Pathology
Streptomyces coelicolor does not cause disease in humans, plants, or animals. Other Streptomyces species, however, are plant pathogens. For example, Streptomyces scabies causes potato scabs, and Streptomyces ipomoeae causes disease in sweet potatoes(17,18).
Edited by Amy Stapp, student of Rachel Larsen at UCSD.
Application to Biotechnology
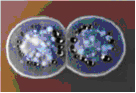
Streptomyces species produce a majority of the antibiotics that have been discovered, so they are very importnat to biotechnology and the development of new antibiotics. Streptomyces coelicolor produces a number of different antibiotics, a few of which will be discussed here. Clorobiocin is an antibiotic that greatly inhibits DNA gyrase. It is not in use pharmacutically at this point, but it may be used as a starting material to make new antibiotics. Production of clorobiocin is controlled in part by the cloY gene, and is similar to a mtbH gene present in Mycobacterium tuberculosis(20). Undecylprodigiosin, also known as Red, is a type of prodiginine produced by Streptomyces coelicolor and is used as anti-tumor agent and an immunosupressant. Production of undecylprodigiosin is controlled by red genes(21). Actinorhodin is another antibiotic produced by Streptomyces coelicolor. This antibiotic is a pH indicator that turns red under acidic conditions and blue under basic conditions, and was very helpful in isolating Streptomyces coelicolor organisms(23). Currently, actinorhodin alone is not used pharmaceutically, but the genes coding for actinorhodin production have been used recombinatorially in other species to form new antibiotic derivatives(22).
Current Research
Research is currently being done to discover the function of different genes in Streptomyces coelicolor since its complete genome has now been sequenced.
Recent research has determined that a group of mtbH-like genes is Streptomyces coelicolor are necessary for some secondary metabolite production. Streptomyces coelicolor has three such genes, one of which is cloY. When all three genes were absent, clorobiocin, an antibiotic, was produced only in very small amounts, but when cloY was restored, clorobiocin was produced at a more significant level. This research also shows that the three genes may be able to "functionally replace each other"(20).
The bld genes are responsible for differentiation in Streptomyces coelicolor. Researchers have determined how the protein BldD interacts in the cell to accomplish this purpose. BldD is a homodimeric, DNA binding protein that has two separately folding subunits. The N terminal half of the protein was determined to be responsible for dimerization and DNA binding. The structure and function of this protein show that BldD may have a very great influence in the develpomental stages of Streptomyces coelicolor(14).
Research is being done to determine how Streptomyces coelicolor use signal transduction pathways to sense changes in their highly variable soil environments, which signals antibiotic and spore production. Link to Researcher's Web-page
The bacterial development of Streptomyces coelicolor is also being studied to determine “the role of specific RNA polymerase holoenzymes controlling development and stress response, global characterisation of spore maturation and germination, cytoskeletal proteins, and chromosome organization during hyphal growth” (9). Link to Researcher's Web-Page
Streptomyces coelicolor is currently the subject of research at the University of Warwick due to its ability to produce prodiginines. These compounds show promise in targeting cancer cells, and a synthetic counterpart to the compound made naturally by Streptomyces coelicolor is in clinical trials as of November 2006(10).
References
(1) "Streptomyces coelicolor A3(2)". NCBI Taxonomy Browser. 29 April 2007. NCBI.
(2) Conn, Jean E. “The Pigment Production of Actinomyces coelicolor and A. violaceus-ruber”. Journal of Bacteriology. 1943. Volume 46. p. 133-149. Link to Article
(3) “From Mapping to Mining the Streptomyces Genome”. John Innes Centre Website. 2001. Link to Article
(4) Thompson, Charles J., Dorris Fink, and Liem D. Nguyen. “Principles of Microbial Alchemy: Insights from the Streptomyces coelicolor Genome Sequence”. Genome Biology 3.7. (2002) Link to Article on PubMed
(5) Kutzner, Hans J and Selman A. Waksman. “Streptomyces coelicolor Muller and Streptomyces violaceoruber Waksman and Curtis, Two Distinctly Different Organisms.” Journal of Bacteriology 78.4 (1959) p. 528-538. Link to Article
(6) Bentley, S.D., K. F. Chater, A.-M. Cerdeño-Tárraga, G. L. Challis , N. R. Thomson, K. D. James, D. E. Harris, M. A. Quail, H. Kieser, D. Harper, A. Bateman, S. Brown, G. Chandra, C. W. Chen, M. Collins, A. Cronin, A. Fraser, A. Goble, J. Hidalgo, T. Hornsby, S. Howarth, C.-H. Huang, T. Kieser, L. Larke, L. Murphy, K. Oliver, S. O'Neil, E. Rabbinowitsch, M.-A. Rajandream, K. Rutherford, S. Rutter, K. Seeger, D. Saunders, S. Sharp, R. Squares, S. Squares, K. Taylor, T. Warren, A. Wietzorrek, J. Woodward, B. G. Barrell, J. Parkhill and D. A. Hopwood. "Complete Genome Sequence of the Model Actinomycete Streptomyces coelicolor A3(2)." Nature. 417. (2002) p. 141-147. Link to Article
(7) Bentley, S. D., S. Brown, L. D. Murphy, D. E. Harris, M. A. Quail, J. Parkhill, B. G. Barrell, J. R. McCormick, R. I. Santamaria, R. Losick, M. Yamasaki, H. Kinashi, C. W. Chen, G. Chandra, D. Jakimowicz, H. M. Kieser, T. Kieser and K. F. Chater. "SPC1, a 356023 bp Linear Plasmid Adapted to the Ecology and Developmental Biology of It's Host, Streptomyces coelicolor." Molecular Microbiology 51.6 (2004) p. 1615-1628.
(8) Haug, Iris, Anke Weissenborn, Dirk Brolle, Stephen Bentley, Tobias Kieser, and Josef Altenbuchner. “Streptomyces Coelicolor A3(2) Plasmid SCP2*: Deductions from the Complete Sequence”. Microbiology 149 (2003). p. 505-513. Link to Article
(9) “Streptomyces: Research.” 30 March 2007. UEA Norwich Website. University of East Anglia. 30 April 2007. http://openwetware.org/wiki/Streptomyces:Research
(10) Stanley, Anna E., Laura J. Walton, Malek Kourdi Zerikly, Christophe Corre and Gregory L. Challis. “Elucidation of the Streptomyces coelicolor pathway to 4-methoxy- 2,29-bipyrrole-5-carboxaldehyde, an intermediate in prodiginine Biosynthesis.” Chemical Communications Articles. (Oct. 2006) RBS Publishing. p. 3981-3983. Link to Article
(11) Borodina, Irina, Preben Krabben, and Jens Nielsen. "Genome-scale Analysis of Streptomyces Coelicolor A3(2) Metabolism". Genome Research. 15 (June 2005) p. 820-828. Link to Article
(12) Keleman, Gabriella H. and Mark J. Buttner. "Initiation of aerial mycelium formation in Streptomyces". Current Opinion in Microbiology. 2.1 (1998) p. 656-662.
(13) Del Sol, Ricardo, Ian Armstrong, Chris Wright, and Paul Dyson. “Characterization of Changes to the Cell Surface during the Life Cycle of Streptomyces coelicolor: Atomic Force Microscopy of Living Cells.” (2007) Journal of Bacteriology. 189.6 p. 2219-2225. Link to Article
(14) Lee, C.J., H.S. Won, J.M. Kim, B.J. Lee, and SO Kang. "Molecular Domain Organization of BldD, an Essential Transcriptional Regulator for Developmental Processes of Streptomyces coelicolor A3(2)." Proteins. 68.1 p. 344-352. Link to Abstract
(15) Viollier, Patrick H., Wolfgang Minas, Glenn E. Dale, Marc Folcher, and Charles J. Thompson. "Role of Acid Metabolism in Streptomyces coelicolor Morphological Differentiation and Antibiotic Biosynthesis." (2001) Journal of Bacteriology. 183.10 p. 3184-3192. Link to Article
(16) van Keulen, G., J. Alderson, J. White and R.G. Sawers. "Nitrate Respiration in the actinomycete Streptomyces coelicolor." (2005) Biochemical Society Transactions. 33.1 p.210 - 212. Link to Article
(17) "Streptomyces scabies". Wellcome Trust Sanger Institute. Link to Website
(18) Zhang, Xiujun, Christopher A. Clark, and Gregg S. Pettis. "Interstrain Inhibition in the Sweet Potato Pathogen Streptomyces ipomoeae: Purification and Characterization of a Highly Specific Bacteriocin and Cloning of Its Structural Gene". (2003) Applied Environmental Microbiology. 69.4 p. 2201-2208. Link to Article
(19) "Streptomyces coelicolor A3(2) Project at Sanger Institute." Entrez Genome Project Website. Link to Website
(20) Wolpert, Manuel, Bertolt Gust, Bernd Kammerer and Lutz Heide. "Effects of deletions of mbtH-like genes on clorobiocin biosynthesis in Streptomyces coelicolor." (2007) Microbiology. 153. p. 1413-1423. Link to Article
(21) White, Janet and Mervyn Bibb. "bldA Dependence of Undecylprodigiosin Production in Streptomyces coelicolor A3(2) Involves a Pathway-Specific Regulatory Cascade." (1999) Journal of Bacteriology. 179.3 p. 627-633. Link to Article
(22) Demain, Arnold L. "Contribution of Genetics to the Production and Discovery of Microbial Pharmaceuticals." (1988) Pure and Applied Chemistry. 60.6 p. 833-836. Link to Article
(23) Ichinose, Koji, Takaaki Taguchi, David J. Bedford, Yutaka Ebizuka, and David A. Hopwood. "Functional Complementation of Pyran Ring Formation in Actinorhodin Biosynthesis in Streptomyces coelicolor A3(2) by Ketoreductase Genes for Granaticin Biosynthesis." (2001) Journal of Bacteriology. 183.10 p. 3247-3250. Link to Article
(24) "Species Specific Metabolic Pathways: Streptomyces ceolicolor." Systems Biology Model Repository. 1 Jun 2007. Link to Website The metabolic pathways listed on this website were taken from the Kyoto Encyclopedia on Genes and Genomes as part of the JST ERATO-SORST Kitano Symbiotic Systems Project.
Edited by Amy Stapp, student of Rachel Larsen and Kit Pogliano
