Epstein-Barr virus (EBV) and EBV-associated lymphomas: Difference between revisions
No edit summary |
|||
| Line 19: | Line 19: | ||
Viral miRNAs are usually hard-to-detect and are 19-23 nt in length <ref name=e>Yang, HJ., Huang, TJ., Yang, CF. et al. Comprehensive profiling of Epstein-Barr virus-encoded miRNA species associated with specific latency types in tumor cells. Virol J 10, 314 (2013). https://doi.org/10.1186/1743-422X-10-314</ref>. They are crucial in maintaining persistent EBV viral infections by promoting tumorigenesis in hosts and down-regulating viral early gene expression. Furthermore, the viral miRNAs are also able to regulate host gene expression at the post-transcriptional level to assist the virus in evading host immune responses. EBV was first found in 2004 to be encoding viral miRNAs, and the to-date 44 mature EBV miRNAs are produced and expressed as two independent transcripts from BHRF1 and BARTs intron prior to splicing <ref name=e/>. Those abundant markers are a hopeful option to be turned into biomarkers for EBV latency types identification in the future. <br><br/> | Viral miRNAs are usually hard-to-detect and are 19-23 nt in length <ref name=e>Yang, HJ., Huang, TJ., Yang, CF. et al. Comprehensive profiling of Epstein-Barr virus-encoded miRNA species associated with specific latency types in tumor cells. Virol J 10, 314 (2013). https://doi.org/10.1186/1743-422X-10-314</ref>. They are crucial in maintaining persistent EBV viral infections by promoting tumorigenesis in hosts and down-regulating viral early gene expression. Furthermore, the viral miRNAs are also able to regulate host gene expression at the post-transcriptional level to assist the virus in evading host immune responses. EBV was first found in 2004 to be encoding viral miRNAs, and the to-date 44 mature EBV miRNAs are produced and expressed as two independent transcripts from BHRF1 and BARTs intron prior to splicing <ref name=e/>. Those abundant markers are a hopeful option to be turned into biomarkers for EBV latency types identification in the future. <br><br/> | ||
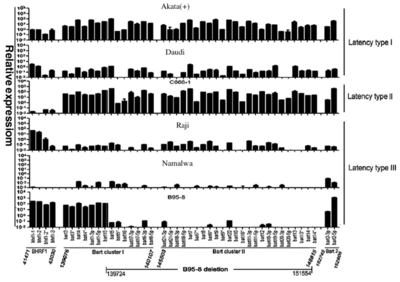
[[Image: fig3A.png|thumb|400px|left|Figure 3. qRT-PCR quantitative analysis of all EBV viral miRNA species' comprehensive expression pattern per latency type. Latency I, II, and III miRNA transcriptome's respective portions are marked below (miR-BHRF1, miR-BAR clusters I/II, miR-BAR2, and B95-8 deletion region). <ref name=e/>]] | [[Image: fig3A.png|thumb|400px|left|Figure 3. qRT-PCR quantitative analysis of all EBV viral miRNA species' comprehensive expression pattern per latency type. Latency I, II, and III miRNA transcriptome's respective portions are marked below (miR-BHRF1, miR-BAR clusters I/II, miR-BAR2, and B95-8 deletion region). <ref name=e/>]] | ||
In | In one recent study, highly sensitive and specific quantitative real-time RT-PCR (qRT-PCR) had been combined with microarray assay for the analysis of patterns of EBV miRNA transcriptomes that are associated with EBV patency types in tumor cells<ref name=e/>. | ||
BHRF1 has been shown to drive chemoresistance and lymphomagenesis by inhibiting multiple cellular pro-apoptotic proteins, thus learning about its expression pattern can help to identify potential therapeutic targets within BHRF1 <ref>Fitzsimmons, L., Cartlidge, R., Chang, C. et al. EBV BCL-2 homolog BHRF1 drives chemoresistance and lymphomagenesis by inhibiting multiple cellular pro-apoptotic proteins. Cell Death Differ 27, 1554–1568 (2020). https://doi.org/10.1038/s41418-019-0435-1</ref>. The general BHRF1 miRNA expression is dependent on the viral latency type as well as the cell’s historical origin. In Figure 3, it can be seen that a nearly 100-fold difference is present between all tested cell lines. Within each latency type and cell lines, BHRF-1 miRNA can still have more than a 50-fold difference despite the fact that those miRNAs were transcribed together as a single transcript. The difference in expression level indicates that the BHRF1 family is latency III dependent as it is almost non-expressing within latency type II and low in expression in latency I <ref name=e/>. | BHRF1 has been shown to drive chemoresistance and lymphomagenesis by inhibiting multiple cellular pro-apoptotic proteins, thus learning about its expression pattern can help to identify potential therapeutic targets within BHRF1 <ref>Fitzsimmons, L., Cartlidge, R., Chang, C. et al. EBV BCL-2 homolog BHRF1 drives chemoresistance and lymphomagenesis by inhibiting multiple cellular pro-apoptotic proteins. Cell Death Differ 27, 1554–1568 (2020). https://doi.org/10.1038/s41418-019-0435-1</ref>. The general BHRF1 miRNA expression is dependent on the viral latency type as well as the cell’s historical origin. In Figure 3, it can be seen that a nearly 100-fold difference is present between all tested cell lines. Within each latency type and cell lines, BHRF-1 miRNA can still have more than a 50-fold difference despite the fact that those miRNAs were transcribed together as a single transcript. The difference in expression level indicates that the BHRF1 family is latency III dependent as it is almost non-expressing within latency type II and low in expression in latency I <ref name=e/>. | ||
Revision as of 02:41, 8 April 2021
By Yangyang Liu (Kenyon '23)
Introduction
Epstein-Barr virus (EBV), formally called Human gammaherpesvirus 4, are a group within the Lymphocryptovirus genus of the Herpesviridae family. EBVise double-stranded DNA viruses that use RNA polymerase for mRNA synthesis based on their negative-strand as the template. EBV Infection occurs through oral transfer of saliva as well as genital secretions and more than 90% of normal adults would gain adaptive immunity after their primary EBV infection. The remaining EBV virus would remain in an asymptomatic latent state for a lifetime within resting B-cells, and a healthy adult with a working immune system would be able to contain the infection with the help of their cytotoxic T cells (CTLs), lymphocyte CD8+ and CD4+, and natural killer (NK) cells [2]. Only a small subset encountering life-threatening diseases due to their unable in maintaining the virus within the latent state. In an uncontrolled situation, EBV-driven lymphoproliferative disorders and lymphomas could develop with the patients. EBV-associated cancers are a common example among the ~15% of all human cancers involving a virus infection [3].
Some of the most well-known illnesses that are caused by EBV are mononucleosis, Burkitt’s lymphoma, Hodgkin’s lymphoma, non-Hodgkin lymphoma, post-transplant lymphoproliferative disease (PTLD), nasopharyngeal carcinoma, and numerous other types of cancer [4]. So far, there is limited success in antivirals development to combat EBV infection. With EBV’s putative role in carcinogenesis, it is important to learn about the mechanisms by which the virus alters host cell characteristics and thus evades the immune system of the hosts. This page investigates how different latency patterns of EBV infection could lead to numerous types of lymphoma, the EBV-associated NK-cell lymphoproliferative diseases (LPD), as well as possible treatment for the specific LPD Chronic active EBV infection (CAEBV).
Identifying Latency Patterns of EBV Infection
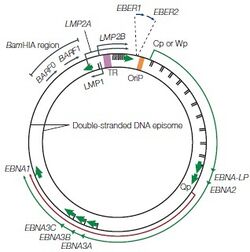
During primary in vivo infection, EBV preferentially infects B-lymphocytes by binding to the CD21 receptor on the cell surface, it will also bind to HLA class II molecules as a co-receptor. In most of the patients, the EBV episome would remain in the latent cycle in resting memory B cells after the primary infection while developing strategies to alter the pattern of the host’s cytotoxic T-lymphocyte (CTL) gene expression. Although there are nearly 100 viral proteins in a regular EBV genome, after infection, the infected resting memory B cells will limit the gene expression to nine viral latent proteins to evade immune recognition. The nine viral latent proteins are composed of six nuclear antigens EBNAs-1, -2, -3a, -3b, -3c, and -LP as well as three latent membrane proteins LMP-1, -2a, -2b. 2 small non-coding RNAs, EBER-1, EBER-2, and BamHI-A rightward transcripts (BART) are also expressed. The expression of nuclear antigens maintains the viral genome, they are also responsible for controlling the expression patterns of the three latent membrane proteins. Different expression combinations of EBNAs and LMPs can lead to different EBV latency programs and the associated lymphomas [6]. Type I latency is only seen in Burkitt’s lymphoma (BL) and selectively express EBNA-1. Type II latency expresses EBNA-1 along with LMP-1 and LMP-2, the common lymphoma for type II latency to be found are Hodgkin’s lymphoma (HL) and peripheral T-cell lymphoma (PTCL). The third and last EBV latency program type III, is commonly found in PTLD and AIDS-related lymphoma. All nine EBV latent proteins are expressed in type III latency [6].
miRNAs for Viral Latency Identification
The EBV latency type classification is still ongoing work, as the traditional classification system stated above is only based on a few molecular markers of EBV latency which introduces limitations to typing of EBV latency types. As different latent infections are associated with different tumors, inaccurate latency identification is an academic and clinical issue. MicroRNAs (miRNAs) species have been found in EBV-infected cells, and a total of 44 EBV miRNA have been reported to be involved in EBV viral latency and tumorigenesis. miRNAs are small non-coding RNAs that broadly regulate gene expression[7].
Viral miRNAs are usually hard-to-detect and are 19-23 nt in length [8]. They are crucial in maintaining persistent EBV viral infections by promoting tumorigenesis in hosts and down-regulating viral early gene expression. Furthermore, the viral miRNAs are also able to regulate host gene expression at the post-transcriptional level to assist the virus in evading host immune responses. EBV was first found in 2004 to be encoding viral miRNAs, and the to-date 44 mature EBV miRNAs are produced and expressed as two independent transcripts from BHRF1 and BARTs intron prior to splicing [8]. Those abundant markers are a hopeful option to be turned into biomarkers for EBV latency types identification in the future.
In one recent study, highly sensitive and specific quantitative real-time RT-PCR (qRT-PCR) had been combined with microarray assay for the analysis of patterns of EBV miRNA transcriptomes that are associated with EBV patency types in tumor cells[8].
BHRF1 has been shown to drive chemoresistance and lymphomagenesis by inhibiting multiple cellular pro-apoptotic proteins, thus learning about its expression pattern can help to identify potential therapeutic targets within BHRF1 [9]. The general BHRF1 miRNA expression is dependent on the viral latency type as well as the cell’s historical origin. In Figure 3, it can be seen that a nearly 100-fold difference is present between all tested cell lines. Within each latency type and cell lines, BHRF-1 miRNA can still have more than a 50-fold difference despite the fact that those miRNAs were transcribed together as a single transcript. The difference in expression level indicates that the BHRF1 family is latency III dependent as it is almost non-expressing within latency type II and low in expression in latency I [8].
The BamHI A rightward transcripts (BART) family, on the other hand, was expressed differently with the three latency types (with the exception of Namalwa cells). As the main cell source was tumors, this suggests that the BART family plays different roles in tumors with different types of latency. The BART family also showed the overall highest amount of expression within the latency type II cell line C666-1, indicating that they may contribute more to the NPC tumorigenesis (which is the origin of tumor cell line C666-1)[8]. Such findings have been shown in previous studies where BARTs are consistently and highly expressed in NPC [10]. Furthermore, the lack of expression within the Namalwa BL cell line could indicate the occasional loss of EBV episomes within lymphoma cells after long-term culture.
Overall, it can be seen that Figure 3 shows evidence indicating the functional role of EBV-encoded miRNA species in showing the expression trends of the EBV viral transcripts within the three latent types. These differentially expressed viral miRNAs are hopeful future molecular biomarkers of latent infection type identification. The BHRF1 family and BART family mentioned in particular could contribute greatly to epithelial tumors and to some lymphocyte tumors latency type differentiation and identification.
Spontaneous Latenty Type Transition Identification Using miRNAs
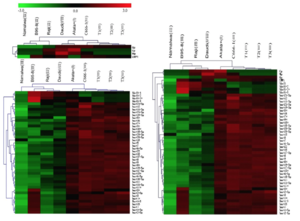
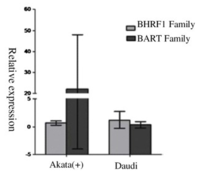
EBV is capable of switching latency types spontaneously in lymphoma cells in vitro. It has been difficult to detect and identify subtle latency transitions due to the lack of specificity and sensitivity of the traditional latency biomarkers. Researchers have tried using traditional biomarkers or miRNA alone, as well as the combination of both to increase the accuracy in detecting latency type switching among tumor cells. As stated in Figure 4, traditional markers alone indicated latency group I for the Burkitt's lymphoma cell line, Daudi [11]. On the other hand, EBV miRNA, as well as the traditional+miRNA combination, grouped the Daudi cells into latency group III.
From the genotypic evaluation of the Daudi cells in comparison to the latent type I cell Akata (+), we are able to evaluate the accuracy of the clustering results from EBV miRNAs (Figure 5). Within Figure 5, it was clear that Daudi’s BHRF1 and BART family relative expression differs from the latent type I cell Akata (+). With the contrasting outcome of Daudi cells EBV viral transcript relative expression, it seems that there was a transition of the Daudi cells from type I to type III latency. It also seems that traditional viral latency biomarkers are an inadequate approach to distinguish the different EBV latency types, whereas the miRNA species that express differentially among latency types may fit the demands better.
EBV-associated rare NK/T-cell Lymphoproliferative Diseases
EBV is a B-lymphotropic virus, so it does not infect the T- and NK-cell lineages during its regular infection cycle in vivo [12]. However, ectopic T and/or NK cell infection does occur on very rare occasions, leading to EBV-associated T and NK-cell lymphoproliferative diseases (EBV+T/NK LPDs) that are characterized by the transformation and proliferation of EBV-infected T or NK cells [13]. The current acknowledged LPDs are classified into six categories, they are chronic active EBV infection (CAEBV) of T- and NK-cell type, systemic EBV-positive T-cell lymphoma of childhood, aggressive NK-cell leukemia (ANKL), extranodal NK/T-cell lymphoma (ENKTL), nasal type, and primary EBV-positive nodal T/NK-cell lymphoma. Here we will focus on the study of CAEBV, since it is one of the LPDs that have a strong ethnic and geographical predilection towards Asians [14].
Chronic Active EBV Infection of T- and NK-cell Type (CAEBV)
Dating back to the first case of CAEBV in 1978, CAEBV has been observed to have the highest case frequencies among children and young adults of Asian origin with no difference in frequency occurring among either sex. There is a roughly equal chance of patients getting CAEBV from either NK or T cell infection, but there are clinical differences between NK and T cell infection. NK cell CAEBV has a tendency of being slowly progressing and result in mild symptoms disregarding the high viral load. The NK cell CAEBV, in contrast, are often rapidly progressive and result in more severe cases [14]. Some of the common symptoms include fever, lymphadenopathy, hepatosplenomegaly, pancytopenia, and persistent hepatitis [16]. It has been observed that patients with CAEBV of T-cell type show a significantly poorer 5-year survival rate of 0.59 than those with CAEBV of NK-cell type (probability of 5-year survival, 0.87 in CAEBV of NK-cell type) [13].
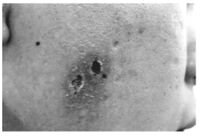
The CAEBV infected cells show a latency type II viral gene expression profile (with the expression of only a few EBV-related genes including EBNA1, LMP1, and LMP2A ) and have the ability to evade immune surveillance while expressing the viral antigens. Although the cause or consequence of this disease is still unclear, a consistently elevated plasma level has been observed in both T and NK cell CAEBV, both with similar pro- and anti-inflammatory cytokine profiles. While the general cytokine profiles of NK and T cell CAEBV are similar, NK cell cases have a significantly higher profile of IL-13. Since IL-13 is involved in IgE responses, NK cell CAEBV patients will have high IgE levels which can lead to hypersensitivity to mosquito bites (Figure 6) [14]. It has also been shown that though NK-type CAEBV has less aggressive clinical behavior in comparison to T-cell CAEBV, CAEBV of NK-types can eventually lead to aggressive NK-cell leukemia or extranodal NK lymphoma, which would have to be treated with chemotherapy [13].
There have been reports that suggest an unusual lytic replication strain of EBV that is impaired for transformation to be the cause of CAEBV. However, the recent case study of CAEBV patients unaffected father indicates that the unusual EBV strain is not the cause of CAEBV disease. The high disease rate within Asians then suggests a role that genetic background could be playing in the disease. The resulting impaired cytotoxic activity of NK or T cells also suggests the possible effect immunodeficiency could have on patients with CAEBV [17]
Interestingly, CAEBV cases that have been documented for non-Asians tend to have different observations in comparison to those observed in Asians. With the main difference being fewer CAEBV cases being T or NK cells but B cells involved, the older mean age of onset, and milder clinical disease symptoms. There is also a progressive loss of B cells and numbers of NK cells within US cases that leads to lethal progressive B-lymphoproliferation or incidental infection due to weakened immune system [14].
Optimal Treatment for CAEBV
The etiology of CAEBV remains unknown, there is also no better current approach to CAEBV optimal treatment other than trying out numerous agents for treatment. Patients without treatment will develop progressive cellular immunodeficiencies, opportunistic infections, as well as multi-organ failure. CAEBV has a poor prognosis and is refractory to treatment such as interferon, antiviral therapy, chemotherapy, as well as intravenous immunoglobulin [19].
The reasoning behind the poor efficacy of antiviral therapy lies mainly in the viral replication cycle. Antiviral therapy acts by inhibiting EBV viral DNA polymerase and therefore inhibiting the replication of EBV during the replication process of its lytic cycle. However, the CAEBV patients’ infected NK or T cells tend to express latent instead of lytic viral gene transcripts. As there is no requirement of viral DNA polymerase for latent EBV, antiviral therapy tends to be ineffective against CAEBV patients [19].
Although the current treatment available are insufficient against treating patients with CAEBV, the main purpose of the treatment is to control the neoplasm and inflammatory disease caused by CAEBV infection. Therefore, the treatment for CAEBV should begin before possible fatal diseases such as HLH or lymphoma develops [20].
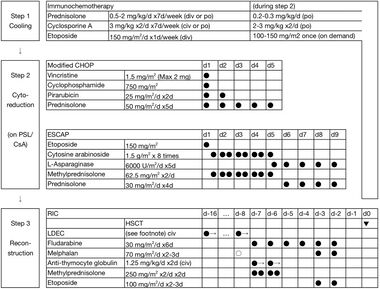
The only known cure for CAEBV is allogenic hematopoietic stem cell transplant (HSCT). And since it could be difficult to diagnose CAEBV on-site, a unified treatment strategy has been developed with HSCT being included in the step 3 reconstruction treatment. There are three steps to the treatment of EBV+T/NK-LPDs (Figure 7). Step one begins with immunochemotherapy and moves on to 2-3 courses of chemotherapy in step two. The final step of CAEBV treatment remains to be HSCT, where cord blood transplantation (CBT) and bone marrow transplantation (BMT) being fitting sources.
The usage of multi-drug chemotherapy in the second step of CAEBV treatment is still up to debate. As one argues that multi-drug chemotherapy could prepare the patient for subsequent allogeneic HSCT by suppressing disease activity, higher rate of donor cells engraftment at HSCT, as well as a low relapse rate after HSCT [19]. Although the benefits have been stated for multi-drug chemotherapy, the real-life application remains complicated. The majority of CAEBV patients died of progression of the disease within the observation periods, which makes it hard for the physicians to prove their hypothesis in practice [19]. While the three-step strategy is applicable to both children and adults, the complexity of nature and small sample sizes for HSCT treatments has slowed down the progress of developing better structured stepwise treatments to yield higher survival rates within CAEBV patients. With the inflammatory symptoms of CABEV being closely related to the outcomes, it is critical for more effective chemotherapy to be developed and incorporated into the three-step treatment [20].
Conclusion
Epstein-Barr virus (EBV) is a member of the herpes virus family, it is an etiological cause of many human lymphocytic and epithelial malignancies, with the most well-known infection being mononucleosis [2]. EBV viral genes are expressed with three latency types. To date, there are 44 EBV-encoded miRNA species found [8]. miRNAs are important in regulating post-transcriptional protein expression, it also plays a physiological role in cell proliferation, differentiation, as well as apoptosis. The EBV miRNA has been found to be representative of a cluster of non-coding latency state biomarkers. It has been found that EBV-coded miRNAs are differentially expressed within EBV-associated tumor cells, and when in comparison to traditional biomarkers miRNAs are able to distinguish between the latent state with higher accuracy [8].
Out of the various diseases EBV is associated with, EBV-associated T/NK cell lymphoproliferative disease (LPDs) are a rare occurrence with NK/T-cell EBV viral infection. Chronic active EBV infection of T and NK-cell type (CAEBV) is one of the most endemic LPDs diseases among East Asia, where there is no sufficient prognosis to date [19]. Thus it is necessary to research the molecular mechanisms of CAEBV development to develop effective treatments or improve the current three-step unified treatment.
References
- ↑ Epstein-Barr virus protein can “switch on” risk genes for autoimmune diseases. National Institutes of Health (NIH). Retrieved 8 April 2021, from https://www.nih.gov/news-events/news-releases/epstein-barr-virus-protein-can-switch-risk-genes-autoimmune-diseases
- ↑ 2.0 2.1 Roschewski, M., & Wilson, W. H. (2012). EBV-associated lymphomas in adults. Best practice & research. Clinical haematology, 25(1), 75–89. https://doi.org/10.1016/j.beha.2012.01.005
- ↑ Amon, W., & Farrell, P. (2005). Reactivation of Epstein-Barr virus from latency. Reviews In Medical Virology, 15(3), 149-156. https://doi.org/10.1002/rmv.456
- ↑ Gequelin, L. C., Riediger, I. N., Nakatani, S. M., Biondo, A. W., & Bonfim, C. M. (2011). Epstein-Barr virus: general factors, virus-related diseases, and measurement of viral load after transplant. Revista brasileira de hematologia e hemoterapia, 33(5), 383–388. https://doi.org/10.5581/1516-8484.20110103
- ↑ Young LS, Rickinson AB. Epstein-Barr virus: 40 years on. Nat Rev Cancer. 2004 Oct;4(10):757-68. doi: 10.1038/nrc1452. PMID: 15510157
- ↑ 6.0 6.1 Roschewski, M., & Wilson, W. H. (2012). EBV-associated lymphomas in adults. Best practice & research. Clinical haematology, 25(1), 75–89. https://doi.org/10.1016/j.beha.2012.01.005
- ↑ Forte, E., & Luftig, M. A. (2011). The role of microRNAs in Epstein-Barr virus latency and lytic reactivation. Microbes and infection, 13(14-15), 1156–1167. https://doi.org/10.1016/j.micinf.2011.07.007
- ↑ 8.0 8.1 8.2 8.3 8.4 8.5 8.6 8.7 8.8 8.9 Yang, HJ., Huang, TJ., Yang, CF. et al. Comprehensive profiling of Epstein-Barr virus-encoded miRNA species associated with specific latency types in tumor cells. Virol J 10, 314 (2013). https://doi.org/10.1186/1743-422X-10-314
- ↑ Fitzsimmons, L., Cartlidge, R., Chang, C. et al. EBV BCL-2 homolog BHRF1 drives chemoresistance and lymphomagenesis by inhibiting multiple cellular pro-apoptotic proteins. Cell Death Differ 27, 1554–1568 (2020). https://doi.org/10.1038/s41418-019-0435-1
- ↑ Marquitz, A. R., & Raab-Traub, N. (2012). The role of miRNAs and EBV BARTs in NPC. Seminars in cancer biology, 22(2), 166–172. https://doi.org/10.1016/j.semcancer.2011.12.001
- ↑ Jones, M. D., Foster, L., Sheedy, T., & Griffin, B. E. (1984). The EB virus genome in Daudi Burkitt's lymphoma cells has a deletion similar to that observed in a non-transforming strain (P3HR-1) of the virus. The EMBO journal, 3(4), 813–821.
- ↑ Shannon-Lowe, C., Rickinson, A. B., & Bell, A. I. (2017). Epstein-Barr virus-associated lymphomas. Philosophical transactions of the Royal Society of London. Series B, Biological sciences, 372(1732), 20160271. https://doi.org/10.1098/rstb.2016.0271
- ↑ 13.0 13.1 13.2 Kim, W., Montes-Mojarro, I., Fend, F., & Quintanilla-Martinez, L. (2019). Epstein-Barr Virus-Associated T and NK-Cell Lymphoproliferative Diseases. Frontiers In Pediatrics, 7. https://doi.org/10.3389/fped.2019.00071
- ↑ 14.0 14.1 14.2 14.3 Shannon-Lowe, C., Rickinson, A. B., & Bell, A. I. (2017). Epstein-Barr virus-associated lymphomas. Philosophical transactions of the Royal Society of London. Series B, Biological sciences, 372(1732), 20160271. https://doi.org/10.1098/rstb.2016.0271
- ↑ Chung, J. S., Shin, H. J., Lee, E. Y., & Cho, G. J. (2003). Hypersensitivity to mosquito bites associated with natural killer cell-derived large granular lymphocyte lymphocytosis: a case report in Korea. The Korean journal of internal medicine, 18(1), 50–52. https://doi.org/10.3904/kjim.2003.18.1.50
- ↑ Kimura, H., & Fujiwara, S. (2019). Overview of EBV-Associated T/NK-Cell Lymphoproliferative Diseases. Frontiers in pediatrics, 6, 417. https://doi.org/10.3389/fped.2018.00417
- ↑ Kimura, H., & Cohen, J. I. (2017). Chronic Active Epstein-Barr Virus Disease. Frontiers in immunology, 8, 1867. https://doi.org/10.3389/fimmu.2017.01867
- ↑ Sawada, A., & Inoue, M. (2018). Hematopoietic Stem Cell Transplantation for the Treatment of Epstein-Barr Virus-Associated T- or NK-Cell Lymphoproliferative Diseases and Associated Disorders. Frontiers in pediatrics, 6, 334. https://doi.org/10.3389/fped.2018.00334
- ↑ 19.0 19.1 19.2 19.3 19.4 Kimura, H., & Cohen, J. I. (2017). Chronic Active Epstein-Barr Virus Disease. Frontiers in immunology, 8, 1867. https://doi.org/10.3389/fimmu.2017.01867
- ↑ 20.0 20.1 Arai, A. (2019). Advances in the Study of Chronic Active Epstein-Barr Virus Infection: Clinical Features Under the 2016 WHO Classification and Mechanisms of Development. Frontiers In Pediatrics, 7. https://doi.org/10.3389/fped.2019.00014
Authored for BIOL 238 Microbiology, taught by Joan Slonczewski, 2021, Kenyon College.
