Darobactin: Difference between revisions
| Line 8: | Line 8: | ||
==Chemical Structure== | ==Chemical Structure== | ||
[[File:Chemical Structure of Darobactin.png|200px|thumb|left|alt text]] | |||
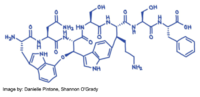
Darobactin, with a chemical formula of C47H55N11O12, is present in the L configuration with two chiral centers and has an unusual structure that has two fused rings that form post-translationally. It is coded by a silent operon and synthesized by the ribosome. Horizontal gene transmission is used to code for Darobactin. The bacteria gathers DNA from the environment and incorporates it into the genome. A unique feature of this antibiotic is the fact that there is a tryptophan-lysine bond between two un-activated carbons. The Darobactin molecule is quite large with a size of 965 Da which is comparable to polymyxin. (Imai et. al. 2019) | Darobactin, with a chemical formula of C47H55N11O12, is present in the L configuration with two chiral centers and has an unusual structure that has two fused rings that form post-translationally. It is coded by a silent operon and synthesized by the ribosome. Horizontal gene transmission is used to code for Darobactin. The bacteria gathers DNA from the environment and incorporates it into the genome. A unique feature of this antibiotic is the fact that there is a tryptophan-lysine bond between two un-activated carbons. The Darobactin molecule is quite large with a size of 965 Da which is comparable to polymyxin. (Imai et. al. 2019) | ||
Revision as of 20:28, 13 May 2021
Introduction
In general, new antibiotics are needed to address the problem of increasing antibiotic resistance. Infections with Gram negative bacteria are particularly challenging to treat. The cellular structure of Gram negative bacteria prevents penetration by many antibiotics and renders many Gram negative pathogens intrinsically antibiotic resistant (Zgurskaya, H. I., Löpez, C. A., & Gnanakaran, S. 2015.) Previously, very few antibiotics have been identified that specifically target Gram negative bacteria. Polymyxin is one example, which functions by disrupting the cell membranes of Gram negative bacteria, destroying their ability to function as osmotic barriers. Rising antibiotic resistance in Gram negative bacteria has threatened the available antibiotics and has highlighted the urgency to discover new antibiotics that specifically target Gram negative pathogens. Therefore, the discovery of Darobactin was of high interest in the medical and biotechnology fields due to the potential implications for the treatment of life threatening infections with Gram negative pathogens.
Origin
The Antimicrobial Discovery Center at Northeastern University, led by Biology professor Dr. Kim Lewis, published their findings on Darobactin in 2019 after two years of research. Darobactin was found in the bacteria Photorhabdus, by Yu Imai, a Northeastern University research associate. The bacteria Photorhabdus was found in the gut of a parasitic worm that is found in soil called a nematode. In the microbiome of the nematode gut, Photorhabdus releases toxins that kill the worm’s prey which allows the worm to digest it.(Candanosa, 2019) Darobactin was discovered in a screen of Photorhabdus isolates to identify compounds produced at a low level that may be encoded within a “silent” biosynthetic gene cluster. This screen identified Photorhabdus khanii HGB1456, and the secreted compound, Darobactin, was found to inhibit the growth of Gram negative bacteria, but not Gram positive bacteria. This drug was tested to treat infections from gram negative infections such as E.coli and Klebsiella. They were able to kill these infections without being toxic to the body, as demonstrated in a mouse infection model. (Imai et. al. 2019)
Chemical Structure
Darobactin, with a chemical formula of C47H55N11O12, is present in the L configuration with two chiral centers and has an unusual structure that has two fused rings that form post-translationally. It is coded by a silent operon and synthesized by the ribosome. Horizontal gene transmission is used to code for Darobactin. The bacteria gathers DNA from the environment and incorporates it into the genome. A unique feature of this antibiotic is the fact that there is a tryptophan-lysine bond between two un-activated carbons. The Darobactin molecule is quite large with a size of 965 Da which is comparable to polymyxin. (Imai et. al. 2019)
Mode of action
As mentioned previously, gram negative bacteria have both inner and outer membranes which makes it difficult for antibiotics to work because of that double layered shield. Darobactin specifically binds to the BamA (β-barrel assembly machine) protein which is an outer membrane protein but works as a tunnel, allowing access into the interspatial space right before the inner membrane. Within this space, there are these nascent outer membrane proteins (OMP) which serve as virulence factors for nutrient scavenging and evasion of host defence mechanisms, making foreign molecules and antibodies very difficult to pass through both membranes The bacteria then inhibits the gram negative cell from absorbing proteins that allow it to make the cell envelope. By restricting this process, the bacteria cannot create and maintain a proper cell envelope resulting in the decay and the death of said gram negative bacteria. In related studies, BamA mutations appear to resist Darobactin, but further Darobactin resistance studies are currently ongoing. With the addition of Darobactin when introduced to E. coli, a well known gram negative bacteria, there was blebbing of the membrane, and eventual swelling and lysis of cells which were promising results that this antibiotic actually did the trick.
Conclusion
The chemical structure of this compound does not inhibit the growth of all gram negative bacteria as of right now, but with this basic discovery and understanding of the mode of action this antibiotic takes, this is a great start in the right direction to have more breakthroughs regarding the creation of antibiotics specifically for gram negative bacteria. This compound can be tweaked in many different ways in order to make it a more broad spectrum antibiotic allowing the same mechanism to take place in regards to the attachment on the BamA protein and the ability to break through both of the membranes. It’s the combination of understanding all the little things that make the big picture easy to comprehend. With this knowledge and all of the experiments that are being done in order to test this compound, there is a very strong possibility in the future Darobactrin will be able to treat many more kinds of gram- negative bacteria.
References
Zgurskaya, H. I., Löpez, C. A., & Gnanakaran, S. (2015). Permeability Barrier of Gram-Negative Cell Envelopes and Approaches To Bypass It. ACS infectious diseases, 1(11), 512–522. https://doi.org/10.1021/acsinfecdis.5b00097
Candanosa, R. M. (2019, November 20). A new antibiotic has been hiding in the gut of a tiny worm. It may be our best weapon against drug-resistant bacteria. News Northeastern A new antibiotic has been hiding in the gut of a tiny worm It may be our best weapon against drug resistant bacteria Comments. https://news.northeastern.edu/2019/11/20/can-darobactin-a-new-antibiotic-found-in-a-tiny-worm-become-our-best-weapon-against-drug-resistant-bacteria/.
Imai, Y., Meyer, K. J., Iinishi, A., Favre-Godal, Q., Green, R., Manuse, S., Caboni, M., Mori, M., Niles, S., Ghiglieri, M., Honrao, C., Ma, X., Guo, J. J., Makriyannis, A., Linares-Otoya, L., Böhringer, N., Wuisan, Z. G., Kaur, H., Wu, R., Mateus, A., … Lewis, K. (2019). A new antibiotic selectively kills Gram-negative pathogens. Nature, 576(7787), 459–464. https://doi.org/10.1038/s41586-019-1791-1
Katsnelson, A. (2019, November 22). Novel antibiotic discovered in the guts of worms. C&EN. https://cen.acs.org/pharmaceuticals/antibiotics/Novel-antibiotic-discovered-guts-worms/97/web/2019/11.
Sousa, M. C. (2019, December 9). New antibiotics target the outer membrane of bacteria. Nature News. https://www.nature.com/articles/d41586-019-03730-x?draft=collection&fbclid=IwAR1G2xiGrA-daXiitJe95nFJRSx5T3TiWWgsv5silORz9_E1PDET1n3uPxc.
Rollauer, S. E., Sooreshjani, M. A., Noinaj, N., & Buchanan, S. K. (2015). Outer membrane protein biogenesis in Gram-negative bacteria. Philosophical transactions of the Royal Society of London. Series B, Biological sciences, 370(1679), 20150023. https://doi.org/10.1098/rstb.2015.0023
Kaur, H., Jakob, R. P., Marzinek, J. K., Green, R., Imai, Y., Bolla, J. R., Agustoni, E., Robinson, C. V., Bond, P. J., Lewis, K., Maier, T., & Hiller, S. (2021). The antibiotic darobactin mimics a β-strand to inhibit outer membrane insertase. Nature, 10.1038/s41586-021-03455-w. Advance online publication. https://doi.org/10.1038/s41586-021-03455-w
Edited by Danielle Pintone and Shannon O'Grady, students of Dr. Charlotte Berkes at Merrimack College