Physarum Polycephalum: Difference between revisions
No edit summary |
No edit summary |
||
| Line 1: | Line 1: | ||
[[Category:Short Pages]] | |||
{{Uncurated}} | {{Uncurated}} | ||
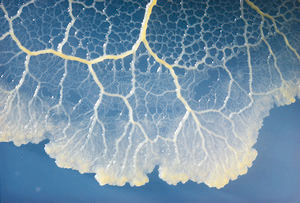
<b><i>P. polycephalum</i></b>, also known as slime mold, is a single celled Eukaryotic organism that grows in the understory or damp, dark places (i.e. rotting material). It is typically yellow in color and consumes other microorganisms, such as fungal spores and bacteria. Slime mold is sensitive to light, although this is what triggers spores to be produced. With a tendency to be very easy to cultivate in the lab, it is used to study mitosis, streaming/cell motility (the movement), and basal forms of intelligence. | <b><i>P. polycephalum</i></b>, also known as slime mold, is a single celled Eukaryotic organism that grows in the understory or damp, dark places (i.e. rotting material). It is typically yellow in color and consumes other microorganisms, such as fungal spores and bacteria. Slime mold is sensitive to light, although this is what triggers spores to be produced. With a tendency to be very easy to cultivate in the lab, it is used to study mitosis, streaming/cell motility (the movement), and basal forms of intelligence. | ||
Latest revision as of 00:11, 7 September 2022
P. polycephalum, also known as slime mold, is a single celled Eukaryotic organism that grows in the understory or damp, dark places (i.e. rotting material). It is typically yellow in color and consumes other microorganisms, such as fungal spores and bacteria. Slime mold is sensitive to light, although this is what triggers spores to be produced. With a tendency to be very easy to cultivate in the lab, it is used to study mitosis, streaming/cell motility (the movement), and basal forms of intelligence.
Classification
Kingdom: Fungi
Phylum: Myxomycota
Class: Myxomycetes
Subclass: Endosporeae
Order: Physarales
Family: Physaraceae
Genus: Physarum
Species: Physarum polycephalum
Note: Some scientists are now classifying this organism in the kingdom Protista because of the way it moves around and feeds.
Life Cycle
1. Plasmodium: the diploid vegetative state that migrates around to find food.
2. Sclerotia: a dormant stage formed in poor growing conditions.
3. Sporulation: triggered by an environmental stress.
4. Sporulation ends with rupture and release of spores.
5. Spores open in ideal environments and release a haploid amoeba.
6. Amoebae:
7. In unfavorable conditions, amoeba form cysts, which are a dormant form.
8. Amoebae form myxoflagellates, or swarm cells, and grow flagella in aqueous solutions.
9. Amoebae can mate and fuse with others that have complementary mating alleles, thus making a new diploid plasmodia.
10. The diploid cells are uninuculeate, but will become multinucleate if cultured.
11. Small plasmodia: can grow large like number 1 if there is a suitable food source.
12. Small plasmodia can be fragmented into microplasmodia.
13. If there is insufficient nutrients, microplasmodia can form Spherules, a dormant form.
P. polycephalum is considered to be a large single celled organism with multiple diploid nuclei, meaning there are no cell walls to distinguish each nuclei. This is when the organism is in its vegetative state. With light stimulation the slime mold begins to retract and can eventually lead to a dormant stage. In this dormancy the slime mold becomes a hardened multinucleated mass which is called sclerotium. It can stay this way for very long periods of time until the environmental conditions are right for it to become vegetative again. When P. polycephalum is in this dormant stage it may begin to go into its reproductive phase. This is where it starts to sporulate (produce spores) and these spores eventually rupture from the cell leaving them exposed for wind and rain to distribute. The spores that are released are flagellated or amoeboid swarm cells in a motile stage and can remain dormant for many years. These swarm cells then come together and fuse, thus creating new diploid plasmodia.
Genome Structure
The entire genome has yet to be sequenced, but there have been some parts that have revealed a good deal about this organism, such as the 26S and 5.8S rRNA genes. The two genes were found to very homologous with regions of E. coli and indicate a link between these, which are eukaryote specific, and the larger prokaryote rRNA gene. Also, while widely considered to be a lower eukaryote, study of aminoacyl-tRNAs has shown that P. polycephalum has patterns of codon recognition that are more like higher plants and animals, and lower fungi.
External Memory
A recent topic of interest and study, this organism has been found to have a form of external memory. As the mold grows and pulsates throughout its environment, it leaves behind a trail of slime marking where it has been (hence the name). When exploring a petri dish, it would not double back to where it has already been before. With no real brain or any way to internalize information, this organism essentially remembers or is reminded of where it has already been when it encounters its own slime again. This type of navigation can be likened to that of the pheromone trails used by ants or bees. Through experimentation, it has been found that slime mold can solve mazes and u-shaped traps (see below) to successfully locate a food source.
References
Gott, J. (n.d.). Genome: Physarum polycephalum. Retrieved from The Genome Institute of Washington University website:http://genome.wustl.edu/genomes/detail/physarum-polycephalum/
Reid R.C, Latty T, Dussutour A, Beekman M. 2012. Slime mold uses an externalized spatial “memory” to navigate in complex environments. PNAS Vol. 109 no. 43: 17490–17494.
Otsuka T, Nomiyama H, Yoshida H, Kukita T, Kuhara S, Sakaki Y. 1983. Complete nucleotide sequence of the 26S rRNA gene of Physarum polycephalum: Its significance in gene history. Proc. NatL Acad. Sci. Vol. 80 no. 11: 3163-3167.
Dussutour A, Latty T, Beekman M, Simpson J. S. 2010. Amoeboid organism solves complex nutritional challenges. Vol. 107 no. 10: 4607–4611.
Hatfield D, Rice M, Hession A. C, Melera W. P. 1982. Journal of Bacteriology. Vol. 151 no.2: 1013-1021
Ridgway B. E, Durham C. H. A. J Cell Biol. 1976 April 1; 69(1): 223–226.