Potyvirus: Difference between revisions
No edit summary |
No edit summary |
||
| Line 11: | Line 11: | ||
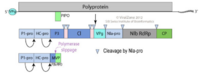
The Potyvirus particle is a long non-enveloped filament that is 680 to 900nm long and 11-20nm wide [11]. The capsin encapsulates a single strand RNA with non-translated 5’-terminal region and 3’-poly-A tail [11][12]. The structure of the virion consists of 10 protein segments: P1-Pro, HC-Pro, P3, CI, NIa, NIb, 6K1, P3N-PIPO, CP, and VPg. The viral protein genome-linked protein (VPg) is located at the 5’ end of the Potyvirus genome [13][14][15]. This VPg is a multifunctional protein that has a role in viral replication and movement [16]. Downstream of VPg, a polyprotein covers the entire open reading frame of the RNA [18][19]. This polyprotein can be cleaved by proteases to form 10 functional and structural proteins. The first protein (P1-pro) is a serine protease that breaks down the polyprotein [20]. The helper component protein (HC-pro) is involved in aphid transmission, transcription initiation in the host cells, and viral RNA silencing suppression [21]. Not much is known about the third protein (P3), but it plays an important role when interacting with the P3N-PIPO movement protein [22]. The cylindrical inclusion protein (CI) is involved in viral replication and cell-to-cell movement, via the formation of conical structures and the interaction with the coat protein [23]. The small nuclear inclusion protein NIa-protease (NIa-pro) is responsible for cleaving the viral polyprotein into functional proteins [24]. The large nuclear inclusion protein NIb-RNA-dependent RNA polymerase (NIb-RdRp) is the center protein for RNA replicase. NIb recruits several host proteins into viral replication complexes (VRCs) [25]. The 6K1 protein, which locates at the 6 kDa position, is known to be a component of the VRC, but function by itself is unknown [26]. The function of the movement protein, P3N-PIPO, is also unknown but has been shown to be required for cell-to-cell movement [22]. The coat protein (CP) is a protein critical for encapsidation, aphid transmission, viral replication, and cell-to-cell movement [27]. | The Potyvirus particle is a long non-enveloped filament that is 680 to 900nm long and 11-20nm wide [11]. The capsin encapsulates a single strand RNA with non-translated 5’-terminal region and 3’-poly-A tail [11][12]. The structure of the virion consists of 10 protein segments: P1-Pro, HC-Pro, P3, CI, NIa, NIb, 6K1, P3N-PIPO, CP, and VPg. The viral protein genome-linked protein (VPg) is located at the 5’ end of the Potyvirus genome [13][14][15]. This VPg is a multifunctional protein that has a role in viral replication and movement [16]. Downstream of VPg, a polyprotein covers the entire open reading frame of the RNA [18][19]. This polyprotein can be cleaved by proteases to form 10 functional and structural proteins. The first protein (P1-pro) is a serine protease that breaks down the polyprotein [20]. The helper component protein (HC-pro) is involved in aphid transmission, transcription initiation in the host cells, and viral RNA silencing suppression [21]. Not much is known about the third protein (P3), but it plays an important role when interacting with the P3N-PIPO movement protein [22]. The cylindrical inclusion protein (CI) is involved in viral replication and cell-to-cell movement, via the formation of conical structures and the interaction with the coat protein [23]. The small nuclear inclusion protein NIa-protease (NIa-pro) is responsible for cleaving the viral polyprotein into functional proteins [24]. The large nuclear inclusion protein NIb-RNA-dependent RNA polymerase (NIb-RdRp) is the center protein for RNA replicase. NIb recruits several host proteins into viral replication complexes (VRCs) [25]. The 6K1 protein, which locates at the 6 kDa position, is known to be a component of the VRC, but function by itself is unknown [26]. The function of the movement protein, P3N-PIPO, is also unknown but has been shown to be required for cell-to-cell movement [22]. The coat protein (CP) is a protein critical for encapsidation, aphid transmission, viral replication, and cell-to-cell movement [27]. | ||
[[File:PotyFigure1.png| | [[File:PotyFigure1.png|200px|thumb|right|'''Figure 1 -''' The gene map and morphology of Potyvirus [50].]] The entire potyvirus consists of one polyprotein that covers the entire RNA stand, with viral protein genome-linked protein (VPg) protein on the 5’ end, and poly-A tail at the 3’ end, with an additional short open reading frame (ORF), called PIPO. The entire RNA codes for the polyprotein with no intron. The polyprotein can be splitted by protease into 10 small protein segments: The first protein (P1-pro), helper component protein (HC-pro), third protein (P3), cylindrical inclusion protein (CI), small nuclear inclusion protein NIa-protease (NIa-pro), large nuclear inclusion protein NIb-RNA-dependent RNA polymerase (NIb-RdRp), and coat protein (CP). The different function of the protein segments leads to replication and movement of the potyvirus. | ||
=5. Metabolic processes= | =5. Metabolic processes= | ||
The Potyvirus enters the host cell via two methods: outside infection, via aphid transmission, and neighboring cell infection, via cell-to-cell movement from an infected cell to its neighboring cell [28]. The viral protein genome-linked protein (VPg) is required to sufficiently transfer the virus RNA of the Potyvirus to enter into the host cell translational machinery without being degraded [29]. Upon entering the host cell, Potyvirus’ polyprotein structure disassembles and releases the genomic RNA, which undergoes translation by the host ribosome and produces a single polyprotein. The polyprotein will then be cleaved by proteases into ten functional proteins to further hijack the host metabolism [30]. | The Potyvirus enters the host cell via two methods: outside infection, via aphid transmission, and neighboring cell infection, via cell-to-cell movement from an infected cell to its neighboring cell [28]. The viral protein genome-linked protein (VPg) is required to sufficiently transfer the virus RNA of the Potyvirus to enter into the host cell translational machinery without being degraded [29]. Upon entering the host cell, Potyvirus’ polyprotein structure disassembles and releases the genomic RNA, which undergoes translation by the host ribosome and produces a single polyprotein. The polyprotein will then be cleaved by proteases into ten functional proteins to further hijack the host metabolism [30]. | ||
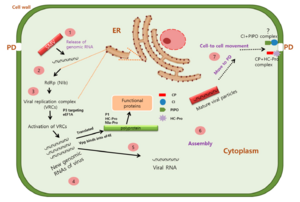
[[File:PotyFigure2.png| | [[File:PotyFigure2.png|300px|thumb|left|'''Figure 2 -''' Replication and movement pathway of soybean mosaic virus(SMV) in genus Potyvirus within the host cell[51]]]. The process of replication is as follows: (1) Release of genomic RNA once a potyvirus enters into the host cell through natural opening like plasmodesmata (PD). (2) translation of potyvirus RNA by ribosome from the large nuclear inclusion protein NIb-RNA-dependent RNA polymerase (NIb-RdRp). (3) NIb-RdRp then hijacked host cell protein eukaryotic translation initiation factor (eIF4E) and isoform (eIF(iso)4E) combined to form the virtual replication complex (VRCs). (4) And active VRCs will continue to replicate new genomic RNAs of the virus. (5) Some of the new viral RNAs will be translated into polyprotein with the initiation by Vpg binding to host eIF4E, and cleaved into functional proteins. (6)Follwoing translation of the polyprotein, virus particles assemble and form the progeny virus. (7) Matured progeny viruses then move to neighbor cells assisted by several proteins. The coat protein(CP) forms a complex with helper component protein (HC-pro) that binds to the progeny virus to protect it from degradation once entered into a new host. Cylindrical inclusion proteins (CI) form complexes with PIPO at the PD, and form a structure that facilitates cell-to-cell movement.|20px]] | ||
The RNA strain of Potyvirus also serves as a template for replication. However, the exact factors that cause virus RNA to shift from translation to replication have yet to be identified. The coat protein (CP) gene is one factor that is associated with the shift of RNA from template for translation to replication [31]. It is known that the NIb-RNA-dependent RNA polymerase (NIb-RdRp) [25], viral protein genome-linked (VPg) [16], cylindrical inclusion protein (CI) [23], and coat protein (CP) [32] of the Potyvirus combined with hijacked host cell protein eukaryotic translation initiation factor (eIF4E) and isoform (eIF(iso)4E) combined form the virtual replication complex (VRCs) which provide functionality for both genome replication and some virion assembly [33]. | The RNA strain of Potyvirus also serves as a template for replication. However, the exact factors that cause virus RNA to shift from translation to replication have yet to be identified. The coat protein (CP) gene is one factor that is associated with the shift of RNA from template for translation to replication [31]. It is known that the NIb-RNA-dependent RNA polymerase (NIb-RdRp) [25], viral protein genome-linked (VPg) [16], cylindrical inclusion protein (CI) [23], and coat protein (CP) [32] of the Potyvirus combined with hijacked host cell protein eukaryotic translation initiation factor (eIF4E) and isoform (eIF(iso)4E) combined form the virtual replication complex (VRCs) which provide functionality for both genome replication and some virion assembly [33]. | ||
Latest revision as of 17:36, 17 October 2024
1. Classification
a. Higher order taxa
Viruses; Riboviria; Orthornavirae; Pisuviricota; Stelpaviricetes; Patatavirales; Potyviridae [1]
2. Description and significance
Potyvirus is a genus of plant virus within the family Potyviridae and is one of the greatest culprits of crop infestation across the entire globe [2]. The word “Poty” derives from the abbreviation of Potato virus Y, which is one of the most well-known species within the genus Potyvirus. The original discovery of Potyvirus can be traced to 7,250 years ago in Southwest Eurasia or North Africa where it diverged from another plant virus of monocotyledons [3]. Potyvirus contains 167 pathogenic species that infect plants, including monocotyledons and dicotyledons [3][7]. The symptoms of the viruses vary, but consist mostly of mosaic symptoms, in which the leaves of the plant are irregularly mottled with different colored streaks or patches, and also leaf malformation. The virulence of Potyvirus ranges from no symptoms to plant death depending on its species and the viral variant [4]. Potyvirus is commonly transmitted via aphids, though some can also be transmitted via seeds, yet the factors that contribute to aphids’ high adaptability are poorly understood [2][8]. Potyvirus can cause a widespread negative impact on the entire agricultural system [4].
3. Genome structure
Currently, only 47 species of Potyvirus have not yet been sequenced. Across all 120 sequenced Potyvirus genomes, most consist of a single-stranded, positive-sense RNA that varies from 9,300 to 10,800nt in length with an average length of 9,799nt [8]. Potyvirus RNA consists of 41.95% G + C nucleotides and encodes 3,125 amino acids, which in turn encodes for a single polyprotein. Some fungal-transmitted Potyviruses have two strands of RNA [9]. 96% of the Potyvirus RNA is the coding region, with only the 3’ poly-A tail being the non-coding segment of the RNA [10]. Using genetic mapping, 48 of the 120 organisms in the genus Potyvirus exhibit higher polyprotein variation that is statistically significant, with the highest genomic variation expressed in the nuclear inclusion protein NIa-protease (NIa-pro) protein-coding region [8].
4. Cell structure
The Potyvirus particle is a long non-enveloped filament that is 680 to 900nm long and 11-20nm wide [11]. The capsin encapsulates a single strand RNA with non-translated 5’-terminal region and 3’-poly-A tail [11][12]. The structure of the virion consists of 10 protein segments: P1-Pro, HC-Pro, P3, CI, NIa, NIb, 6K1, P3N-PIPO, CP, and VPg. The viral protein genome-linked protein (VPg) is located at the 5’ end of the Potyvirus genome [13][14][15]. This VPg is a multifunctional protein that has a role in viral replication and movement [16]. Downstream of VPg, a polyprotein covers the entire open reading frame of the RNA [18][19]. This polyprotein can be cleaved by proteases to form 10 functional and structural proteins. The first protein (P1-pro) is a serine protease that breaks down the polyprotein [20]. The helper component protein (HC-pro) is involved in aphid transmission, transcription initiation in the host cells, and viral RNA silencing suppression [21]. Not much is known about the third protein (P3), but it plays an important role when interacting with the P3N-PIPO movement protein [22]. The cylindrical inclusion protein (CI) is involved in viral replication and cell-to-cell movement, via the formation of conical structures and the interaction with the coat protein [23]. The small nuclear inclusion protein NIa-protease (NIa-pro) is responsible for cleaving the viral polyprotein into functional proteins [24]. The large nuclear inclusion protein NIb-RNA-dependent RNA polymerase (NIb-RdRp) is the center protein for RNA replicase. NIb recruits several host proteins into viral replication complexes (VRCs) [25]. The 6K1 protein, which locates at the 6 kDa position, is known to be a component of the VRC, but function by itself is unknown [26]. The function of the movement protein, P3N-PIPO, is also unknown but has been shown to be required for cell-to-cell movement [22]. The coat protein (CP) is a protein critical for encapsidation, aphid transmission, viral replication, and cell-to-cell movement [27].
The entire potyvirus consists of one polyprotein that covers the entire RNA stand, with viral protein genome-linked protein (VPg) protein on the 5’ end, and poly-A tail at the 3’ end, with an additional short open reading frame (ORF), called PIPO. The entire RNA codes for the polyprotein with no intron. The polyprotein can be splitted by protease into 10 small protein segments: The first protein (P1-pro), helper component protein (HC-pro), third protein (P3), cylindrical inclusion protein (CI), small nuclear inclusion protein NIa-protease (NIa-pro), large nuclear inclusion protein NIb-RNA-dependent RNA polymerase (NIb-RdRp), and coat protein (CP). The different function of the protein segments leads to replication and movement of the potyvirus.
5. Metabolic processes
The Potyvirus enters the host cell via two methods: outside infection, via aphid transmission, and neighboring cell infection, via cell-to-cell movement from an infected cell to its neighboring cell [28]. The viral protein genome-linked protein (VPg) is required to sufficiently transfer the virus RNA of the Potyvirus to enter into the host cell translational machinery without being degraded [29]. Upon entering the host cell, Potyvirus’ polyprotein structure disassembles and releases the genomic RNA, which undergoes translation by the host ribosome and produces a single polyprotein. The polyprotein will then be cleaved by proteases into ten functional proteins to further hijack the host metabolism [30].
. The process of replication is as follows: (1) Release of genomic RNA once a potyvirus enters into the host cell through natural opening like plasmodesmata (PD). (2) translation of potyvirus RNA by ribosome from the large nuclear inclusion protein NIb-RNA-dependent RNA polymerase (NIb-RdRp). (3) NIb-RdRp then hijacked host cell protein eukaryotic translation initiation factor (eIF4E) and isoform (eIF(iso)4E) combined to form the virtual replication complex (VRCs). (4) And active VRCs will continue to replicate new genomic RNAs of the virus. (5) Some of the new viral RNAs will be translated into polyprotein with the initiation by Vpg binding to host eIF4E, and cleaved into functional proteins. (6)Follwoing translation of the polyprotein, virus particles assemble and form the progeny virus. (7) Matured progeny viruses then move to neighbor cells assisted by several proteins. The coat protein(CP) forms a complex with helper component protein (HC-pro) that binds to the progeny virus to protect it from degradation once entered into a new host. Cylindrical inclusion proteins (CI) form complexes with PIPO at the PD, and form a structure that facilitates cell-to-cell movement.|20px]]
The RNA strain of Potyvirus also serves as a template for replication. However, the exact factors that cause virus RNA to shift from translation to replication have yet to be identified. The coat protein (CP) gene is one factor that is associated with the shift of RNA from template for translation to replication [31]. It is known that the NIb-RNA-dependent RNA polymerase (NIb-RdRp) [25], viral protein genome-linked (VPg) [16], cylindrical inclusion protein (CI) [23], and coat protein (CP) [32] of the Potyvirus combined with hijacked host cell protein eukaryotic translation initiation factor (eIF4E) and isoform (eIF(iso)4E) combined form the virtual replication complex (VRCs) which provide functionality for both genome replication and some virion assembly [33].
Cell-to-cell movement of the viral RNA is facilitated by viral proteins to travel across the plasmodesmata, which connects the cytoplasm and ER of the host plant cell to neighboring cells. The three proteins involved in this process are: movement protein P3N-PIPO, cylindrical inclusion protein (CI), and coat protein (CP) [34][35][36].
One unique feature of Potyvirus is the pinwheeled-like structures called the cylindrical inclusion proteins. These proteins are involved in replication processes during viral infection [32]. Their interactions with the coat proteins also play a direct role in the intercellular migration of the viral genome, whereby the proteins can induce a conical structure that facilitates their ability to pierce through cell walls and associate with other neighboring cells [37].
The helper component proteinase (HC-Pro) is another crucial protein found within the polyprotein complex that is synthesized from the viral RNA encoded by Potyvirus [38]. It is one of the most well-studied proteins in the genus given its profound roles in viral transmission, viral maturation, and RNA silencing suppression [38]. One specific function of HC-Pro is its role in protein synthesis via interactions with two translation initiation factors, called eIF4E and eIF(iso)4E, without interacting with VPg. HC-Pro has a positive effect on translation activity, which is crucial in viral maturation on a larger scale [39]. Another notable function of HCPro is is its ability to demethylate and activate the auxin biosynthesis genes in plants, which induces an overexpression of the genes and subsequently interferes with the auxin signaling pathways that induce the defense mechanism against viral pathogens in plants [40]. The HCPro and the CP proteins also play a direct role in attaching to the stylet of aphids and facilitating the transmission of the virus to other host plants [5].
6. Ecology
There are 167 species of Potyvirus distributed around the world [8]. The US currently has 81 species of the virus, with China having 63, Australia having 62, and France having 49 [5]. Each species of Potyvirus targets a specific range of plant hosts. These hosts can range from domesticated crops such as potatoes or tobacco, targeted by Potato virus Y, to wild plants such as Conium maculatum and Ruta montana, targeted by Poison hemlock virus Y and Mediterranean ruda virus, respectively, though the majority of infestations are still found in agricultural crops [41][42][43].
Potyvirus can be transmitted through more than 200 species of aphids [5]. The transmission is in a non-persistent manner, indicating the virus only resides within the insect vectors (insect stylets) for a short period of time relative to persistent transmission [6]. Although there are over 200 known species of aphids involved in this transmission process, the majority of the Potyvirus species are transmitted by only a few species of aphids. These few aphid species are mostly polyphagous, meaning they feed on various families of plants [5]. Their wide range of food sources explain why they have a greater range of transmission than monophagous aphids that feed on only one genus or family of plants. For example, M. persicae, which is polyphagous, is responsible for transmitting 53.4% of all the Potyvirus species while L. erysimi, which is monophagous, transmits only 4.5% of the species [5]. Nevertheless, monophagous aphid species are still capable of having high transmission efficiency regardless of transmission range. For instance, M. persicae is more efficient at transmitting Potato Virus Y to Capsicum annuum, whereas L. erysimi is more efficient at transmitting Turnip mosaic virus to B. rapa [5].
7. Pathology and Epidemiology
Potyvirus is a widespread plant virus responsible for a significant portion of crop losses among the agricultural industry [2]. Aphids act as vectors that transport the viral RNA of Potyviruses from one plant to another. Aphids can acquire the virion within seconds or minutes [5]. Once the virion is acquired, the aphid retains it transiently, during which it can pass on the virion to other nearby host plants [5].
Common phenotypic symptoms of Potyvirus include stunted growth, reduction in tuberous root, and leaf deformation (or mosaics) [44][45]. These symptoms, if left untreated, can result in significant crop loss, as highlighted by the past pandemic of the Sweet potato feathery mottle virus in the Democratic Republic of Congo. The crop loss became so devastating that the entire agricultural industry of sweet potatoes ceased to exist for a period of time due to its unprofitability [45]. Many current countermeasures have hitherto shown to be effective in targeting both Potyvirus and the aphids. For example, selective breeding of viral-resistant crops, amputation of infected plant bodies, or the removal of the entire infected host plant are all effective in stopping the spread of Potato virus Y, Plum pox virus, and Lettuce mosaic virus [5]. On the other hand, the use of physical barriers, mineral oil spraying, and reflective mulches have proven to be effective in repelling aphids. But given the complexity in the transmission and the infection of the virus, the strategy of integrating multiple countermeasures together is often required to effectively mitigate the damage of widespread crop infection [5].
8. Current Research
Due to the substantial economic loss caused by the Potyvirus, the emergence of improved antiviral strategies against Potyvirus infection is essential. One recent research has put focus on the identification of conserved miRNAs within the host plants [46]. The use of miRNAs based antiviral resistance strategies has been successfully applied against the animal virus but not on plant viruses. The miR168 and miR162 genes were found to be conserved in many plants, which can potentially be used to help design broad-spectrum antiviral strategies [46].
Thermotherapy and chemotherapy were also considered for plant virus elimination but studies showed these therapies alone are not efficient in viral elimination due to the low surviving rate and reproduction rate. A recent study explores the possible combination of thermotherapy and explant size in hope to improve the efficiency of eliminating Potyvirus and Carlavirus from infected shallot in Indonesia [47]. The results show that the frequency of viral infection is lowered when explant size at 37 °C is decreased. Although the elimination efficiency does show signs of improvements, the reproduction rate was not meeting the expectancy and more research can be conducted for future improvement [47].
Future directions of Potyvirus study include the multi-omics approaches such as RNAi and CRISPR-based gene editing. These techniques are used to develop efficient and sustainable virus vector management strategies. RNAi plays a role in plant virus defense by degrading target RNA molecules at transcriptional or post-transcriptional levels [9]. A recent study found that dsRNAs targeting NIb or CP protect N. benthamiana (relative of tobacco) and cowpea plants from aphid-mediated transmission [48]. This study provides the possibility of using RNAi for crop protection. CRISPR gene editing can be used to develop tools that genetically modify vectores’ transmission cycle to achieve plant genetic improvement and protection within short times. A recent study has successfully applied CRISPR/Cas13a as the tool to trigger TuMV resistance in N. benthamiana with specific virus interference that knocks down the HC-Pro protein to block the aphid transmission of Potyviruses [49].
9. Authorship Statement
Zhiyue Wu wrote the sections of Classification and Current Research. Duoxi Xian wrote the section of the Introduction and edited the reference. Shiyu Zhang wrote the sections of Genome Structure, Cell Structure, and Metabolic Process. Chun Lin wrote the sections on Ecology and Pathology.
Edited by Zhiyue Wu, Duoxi Xian, Shiyu Zhang, and Chun Lin, student of Jennifer Bhatnagar for BI 311 General Microbiology, 2021, Boston University.
10. References
[4] Abd El-Aziz MH. 2020. The Importance of Potato virus Y Potyvirus. J Plant Sci Phytopathol. 4: 009-015.
[5] Gadhave, K. R., Gautam, S., Rasmussen, D. A., & Srinivasan, R. 2020. Aphid Transmission of Potyvirus: The Largest Plant-Infecting RNA Virus Genus. Viruses, 12(7):773.
[6] Shi, X., Zhang, Z., Zhang, C., Zhou, X., Zhang, D. and Liu, Y. 2021. The molecular mechanism of efficient transmission of plant viruses in variable virus–vector–plant interactions. Horticultural Plant Journal, 7(6), pp.501-508.
[7] Wylie, S. J., Adams, M., Chalam, C., Kreuze, J., López-Moya, J. J., Ohshima, K., Praveen, S., Rabenstein, F., Stenger, D., Wang, A., Zerbini, F. M., & Ictv Report Consortium. 2017. ICTV Virus Taxonomy Profile: Potyviridae. The Journal of general virology, 98(3), 352–354.
[8] Nigam, D., LaTourrette, K., Souza, P., & Garcia-Ruiz, H. 2019. Genome-Wide Variation in Potyviruses. Frontiers in plant science, 10:1439.
[9] Shukla, D.D.; Frcnkel, M.J.; Ward, C.W. 1991. Structure and function of the potyvirus genome with special reference to the coat protein coding region. Canadian Journal of Plant Pathology, 13(2), 178–191.
[10] Frenkel MJ, Jilka JM, Shukla DD, Ward CW. 1992. Differentiation of potyviruses and their strains by hybridization with the 3' non-coding region of the viral genome. J Virol Methods. 36(1):51-62.
[11] Dougherty, W.G. and Carrington, J.C. 1988. Expression and function of potyviral gene products. Annu. Rev. Phytopathol., 26: 123-143.
[12] Van der Vlugt, R., Allefs, S., De Haan, P. and Goldbach, R. 1989. Nucleotide sequence of the 3’-terminal region of potato virus YN RNA. J. Gen. Virol., 70: 229-233.
[13] Hari, V. 1981. The RNA of tobacco etch virus further characterization and detection of protein linked to RNA. Virology 112:391-399.
[14] Siaw, M.F.E., M. Shahabuddin, S. Ballard, J.G. Shaw and R.E. Rhoads. 1985. Identification of a protein evolutionary linked to the 5'-terminus of tobacco vein mottling virus RNA, Virology. 142:134-143.
[15] Riechmann, J.L., S. Lain, and J.A. Garcia. 1989. The genome linked protein and 5'RNA sequence of plum pox potyvirus. Journal Gen Virol. 70:2785-2789.
[16] Rajamäki ML, Valkonen JP. 2002. Viral genome-linked protein (VPg) controls accumulation and phloem-loading of a potyvirus in inoculated potato leaves. Mol Plant Microbe Interact. 15(2):138-49.
[17] Dougherty, W.G.. and J.C. Carrington. (1988). Expression and lunelion of potyviral gene products. Annu. Rev. Phytopathol. 26:123-143.
[18] Hari, V., A. Siegel, C. Rozek, and VV.E. Timberlake. 1979. The RNA of tobacco etch virus contains poly(A) Virology 92:568-571.
[19] Hu, SF., Wei, WL., Hong, SF. et al. 2020. Investigation of the effects of P1 on HC-pro-mediated gene silencing suppression through genetics and omics approaches. Bot Stud 61, 22
[20] Plisson C, Drucker M, Blanc S, German-Retana S, Le Gall O, Thomas D, Bron P. 2003. Structural characterization of HC-Pro, a plant virus multifunctional protein. J Biol Chem. 278(26):23753-61.
[21] Xiaoyan C, Hoda Y, Guanwei W, Xiaoyun W, Xin C, Greg T, Aiming W. 2017. The C-terminal region of the Turnip mosaic virus P3 protein is essential for viral infection via targeting P3 to the viral replication complex. Virology, 2017; 510:147-155.
[22] Deng P, Wu Z, Wang A. 2015. The multifunctional protein CI of potyviruses plays interlinked and distinct roles in viral genome replication and intercellular movement. Virol J. 2015;12:141.
[23] Gong YN, Tang RQ, Zhang Y, et al. 2020. The NIa-Protease Protein Encoded by the Pepper Mottle Virus Is a Pathogenicity Determinant and Releases DNA Methylation of Nicotiana benthamiana. Front Microbiol. 11:102.
[24] Shen W, Shi Y, Dai Z, Wang A. 2020. The RNA-Dependent RNA Polymerase NIb of Potyviruses Plays Multifunctional, Contrasting Roles during Viral Infection. 12(1):77.
[25] Hong, XY., Chen, J., Shi, YH. et al. 2007. The ‘6K1’ protein of a strain of Soybean mosaic virus localizes to the cell periphery. Arch Virol 152, 1547–1551.
[26] Yan, ZY., Xu, XJ., Fang, L. et al. 2021. Multiple aromatic amino acids are involved in potyvirus movement by forming π-stackings to maintain coat protein accumulation. Phytopathol Res 3, 10.
[27] Kristiina, M., Anders, H. 2014. Intracellular coordination of potyviral RNA functions in infection. Front. Plant Sci. 2014.00110
[28] Oruetxebarria I., Guo D., Merits A., Mäkinen K., Saarma M, Valkonen J. P. T. 2001. Identification of the genome-linked protein in virions of Potato virus A, with comparison to other members in genus Potyvirus. Virus Res. 73:103–112.
[29] Rajamäki M., Mäki-Valkama T., Mäkinen K, Valkonen J. P. T. 2004. “Infection with potyviruses,” in Plant–Pathogen Interactions ed. Talbot N.
[30] Mahajan S., Dolja V. V., Carrington J. C. 1996. Roles of the sequence encoding tobacco etch virus capsid protein in genome amplification: requirements for the translation process and a cis-active element. J. Virol. 70:4370–4379.
[31] Sorel, M., Garcia, J. A., & German-Retana, S. 2014. The Potyviridae cylindrical inclusion helicase: a key multipartner and multifunctional protein. Molecular plant-microbe interactions : MPMI, 27(3):215–226.
[32] Kekarainen T., Savilahti H, Valkonen J. P. T. 2002. Functional genomics on Potato virus A: a virus genome-wide map of sites essential for virus propagation. Genome Res. 12:584–594.
[33] Dolja V. V., Haldeman R., Robertson N. L., Dougherty W. G., Carrington J. C. 1994. Distinct functions of capsid protein in assembly and movement of tobacco etch Potyvirus in plants. EMBO J. 13:1482–1491.
[34] Carrington J. C., Jensen P. E., Schaad M. C. 1998. Genetic evidence for an essential role for Potyvirus CI protein in cell-to-cell movement. Plant J. 14:393–400.
[35] Wei T., Zhang C., Hong J., Xiong R., Kasschau K. D., Zhou X., et al. 2010. Formation of complexes at plasmodesmata for Potyvirus intercellular movement is mediated by the viral protein P3N-PIPO. PLoS Pathog.
[36] Rodríguez-Cerezo, E., Findlay, K., Shaw, J. G., Lomonossoff, G. P., Qiu, S. G., Linstead, P., Shanks, M., & Risco, C. 1997. The coat and cylindrical inclusion proteins of a potyvirus are associated with connections between plant cells. Virology, 236(2): 296–306.
[37] Valli, A. A., Gallo, A., Rodamilans, B., López-Moya, J. J., & García, J. A. 2018. The HCPro from the Potyviridae family: an enviable multitasking Helper Component that every virus would like to have. Molecular plant pathology, 19(3):744–763.
[38] Ala-Poikela M, Goytia E, Haikonen T, Rajamäki ML, Valkonen JP. 2011. Helper component proteinase of the genus Potyvirus is an interaction partner of translation initiation factors eIF(iso)4E and eIF4E and contains a 4E binding motif. J Virol. 85(13):6784-6794.
[39] Yang, L., Meng, D., Wang, Y., Wu, Y., Lang, C., Jin, T., & Zhou, X. 2020. The viral suppressor HCPro decreases DNA methylation and activates auxin biosynthesis genes. Virology 546:133–140.
[40] Mao, Y., Sun, X., Shen, J., Gao, F., Qiu, G., Wang, T., Nie, X., Zhang, W., Gao, Y., & Bai, Y. 2019. Molecular Evolutionary Analysis of Potato Virus Y Infecting Potato Based on the VPg Gene. Frontiers in microbiology, 10:1708.
[41] Nury, S., Hosseini, A., Gibbs, A.J. et al. 2020. Poison hemlock virus Y (PHVY), a novel potyvirus from Iranian Conium maculatum (Apiaceae). Australasian Plant Pathol. 49:119–126.
[42] Rodríguez-Nevado C, Montes N and Pagán I. 2017. Ecological Factors Affecting Infection Risk and Population Genetic Diversity of a Novel Potyvirus in Its Native Wild Ecosystem. Front. Plant Sci. 8:1958.
[43] Coutts, B. A., Kehoe, M. A., Webster, C. G., Wylie, S. J., & Jones, R. A. 2011. Indigenous and introduced potyviruses of legumes and Passiflora spp. from Australia: biological properties and comparison of coat protein nucleotide sequences. Archives of virology, 156(10):1757–1774.
[44] Jones R. 2021. Global Plant Virus Disease Pandemics and Epidemics. Plants (Basel, Switzerland), 10(2), 233.
[45] Sankaranarayanan, R., Palani, S.N., Tamilmaran, N. et al. 2021. Novel approaches on identification of conserved miRNAs for broad-spectrum Potyvirus control measures. Mol Biol Rep 48: 2377–2388.
[46] Putri, P.H., Hidayat, S.H., & Dinarty D. 2019. Elimination of Potyvirus and Carlavirus from Infected shallot bulbs. Environment Complete 21(1):42-46.
[47] Worrall E.A., Bravo-Cazar A., Nilon A.T., Fletcher S.J., Robinson K.E., et al. 2019. Exogenous application of RNAi-inducing double-stranded RNA inhibits aphid-mediated transmission of a plant virus. Front Plant Sci. (10) :265.
[48] Aman R., Ali Z., Butt H., Mahas A., Aljedaani F., et al. 2018. RNA virus interference via CRISPR/Cas13a system in plants. Genome Biol. (19) :1.
[49] SIB Swiss Institute of Bioinformatics, Potyvirus Genome, ViralZone. 2012. Accessed December 6th, 2021. https://viralzone.expasy.org/50?outline=all_by_species
[50] Widyasari K, Alazem M, Kim K-H. 2020. Soybean Resistance to Soybean Mosaic Virus. Plants. 9(2):219.