Arthroderma benhamiae: Difference between revisions
| Line 51: | Line 51: | ||
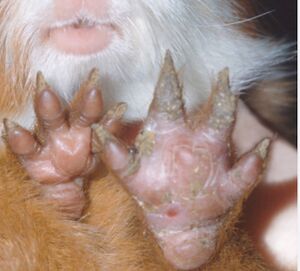
[[Image:GuineaFeet.jpg|thumb|300px|right|Affected feet of Guinea Pig. Image credit:https://www.sciencedirect.com/topics/pharmacology-toxicology-and-pharmaceutical-science/arthroderma-benhamiae ]] | [[Image:GuineaFeet.jpg|thumb|300px|right|Affected feet of Guinea Pig. Image credit:https://www.sciencedirect.com/topics/pharmacology-toxicology-and-pharmaceutical-science/arthroderma-benhamiae ]] | ||
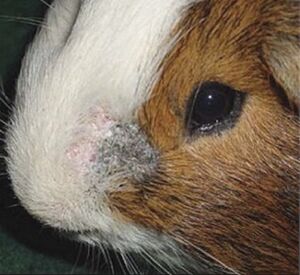
[[Image:Guinea.jpg|thumb|300px|right|Arthroderma lesion. Image credit: https://www.sciencedirect.com/topics/pharmacology-toxicology-and-pharmaceutical-science/arthroderma-benhamiae]] | |||
==References== | ==References== | ||
Revision as of 17:50, 9 December 2024
Classification
Domain: Eukaryota ; Phylum: Fungi ; Class Dikayrya ; Order: ascomycota ; family: Arthroderma
Species
Genus species
Arthroderma benhamiae
|
NCBI: [1] |
Description and Significance
Arthroderma benhamiae is a dermatophyte fungus morph that belongs to the Trichophyton mentagrophytes species. The guinea pig is considered it's natural host as it is primarily carried by guinea pigs and small rodents with the highest infection rate. The fungus is characterized by inflammatory skin lesions that are raised and erythematous and crusty. Infection is usually limited to layers of skin, hair and nails in keratinized hosts. This organism is important to study as dermatophytoses are the most common fungal infections and their primary carriers are commonly kept as household pets. This can increase the infection rates in humans who are handling these pets in close contact. The treatment for a mild infection can include topical treatment, but can be more extensive in some cases where oral antifungals are necessary.
Genome Structure
The genomes of these dermatophytes are extremely small ranging from 22.5 to 24 Mb and are highly collinear. A. benhamiae genome yields 7,405 protein-encoding genes. The genomes in these dermatophytes have been found to be rich with genes encoding secreted proteases where as genes encoding enzymes for sugar metabolism were not present at all. Those genes are particularly important for plant cell wall breakdown which isn't important in our fungus since it primarily infects animal and human skin. A. benhamiae typically has between 8-20 chromosomes.
Cell Structure, Metabolism and Life Cycle
Arthroderma Benhamiae has been studied to use certain biosyntheses of pigments for growth and metabolism. Based on some evidence collected, they connected the pigments to playing an active role in respiration, specifically in the electron transport chain. According to a series of pigment extraction, purification and characterization methods, they found that nitrogen containing amino acid pigments supported growth. They also found that growth was inhibited on the sulfur containing amino acids.
Nitrogen acquisition from the host is imperative for this pathogenic fungus to survive, grow and persist. The dermophytes have a high degree of metabolic flexibility. In the absence of their preferred nitrogen source, they will use amino acids or cetain proteins. To that point, it has been studied that extracellular proteases aid in metabolism of the fungal pathogenic dermophytes like our organism. Acting as a key virulence attribute to the fungus to break down the keratin containing tissues. Their genomes show enrichment of these protease genes which are expressed typically only when in growth on keratin in vitro. This is expected as when they are infecting a host, those specialized genes are up regulated to assist in the degradation of the keratin.
Infection of Arthroderma benhamiae can begin within 2-6 hours of contact; within 24 hours a fungal invasion takes place, causing damage to the keratin network. After 7 days’ incubation period, hyphae form asexual spores that complete the life cycle. Lesions on affected skin start to form between 1-3 weeks after exposure.
Ecology and Pathogenesis
The habitat of our microbe is on the skin of animals and humans! Its normally found on small rodents and therefore gets its opportunity to transfer to humans. It breaks down keratinized tissues and colonizes in the tissue. In humans it normally causes skin infections such as ringworm or tinea, which is athletes foot, jock itch, or scalp infections. But, when the fungus stays on animals it causes only dermatophytosis, and it does not infect plants because they do not have the keratin necessary to sustain their metabolism. In humans this can lead them to have red itchy skin, and circular rashes with raised edges, while in animals, it causes fur loss, redness, and itchy lesions on skin.
Its virulence factors include keratinase enzymes to help digest keratin and allow for invasion. Proteases and lipases help break down the hots proteins and lipids to help fuel the organism. It also has adhesion factors and cell-wall components to help stay on organisms and protect itself.
It can help contribute to the environment by controlling amounts of keratinous debris and recycling it. Its biochemical significance lies in the decomposition of keratin, and helps to recycle it back into the ecosystem so other organisms can use it.
References
1. GHANI HM. Physiological, Chemical, And Genetic Study Of Pigment Production By Arthroderma Benhamiae. [Order No. 7309154]. The University of Oklahoma; 1972.
2. Ene IV, Brunke S, Brown AJ, Hube B. Metabolism in fungal pathogenesis. Cold Spring Harb Perspect Med. 2014;4(12):a019695. Published 2014 Sep 4. doi:10.1101/cshperspect.a019695
3. Tran VD, De Coi N, Feuermann M, et al. RNA Sequencing-Based Genome Reannotation of the Dermatophyte Arthroderma benhamiae and Characterization of Its Secretome and Whole Gene Expression Profile during Infection. mSystems. 2016;1(4):e00036-16. Published 2016 Aug 2. doi:10.1128/mSystems.00036-16
4. Martín-Peñaranda T, Lera Imbuluzqueta JM, Alkorta Gurrutxaga M. Arthroderma benhamiae en pacientes con cobayas. An Pediatr (Barc). 2019;90:51–52.
Author
Page authored by Kathleen Bessette, Cameron Welker, Nicole Buzaki, & Alex Nice, students of Prof. Bradley Tolar at UNC Wilmington.

