Corynebacterium: Difference between revisions
No edit summary |
No edit summary |
||
| Line 1: | Line 1: | ||
{| | {| | ||
| height="221" bgcolor="#FFDF95" | | | height="221" bgcolor="#FFDF95" | | ||
'''NCBI:<br />[http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Info&id=1716&lvl=3&lin=f&keep=1&srchmode=1&unlock Taxonomy]<br />Genome:<br />'''[http://www.ncbi.nlm.nih.gov/genomes/framik.cgi?db=Genome&gi=375 Cornyebacterium diphtheriae]''' | '''NCBI:<br />[http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Info&id=1716&lvl=3&lin=f&keep=1&srchmode=1&unlock Taxonomy]<br />Genome:<br />''''[http://www.ncbi.nlm.nih.gov/genomes/framik.cgi?db=Genome&gi=375 Cornyebacterium diphtheriae]''' | ||
'''[http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?id=257309 <br />]'''[http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?id=257309 C]'''[http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?id=257309 '''orynebacterium efficiens ''YS-31<br />]'''[http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?id=257309 '''Corynebacterum glutamicum'' ATCC 13032]''' | ''''[http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?id=257309 <br />]'''[http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?id=257309 C]'''[http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?id=257309 '''orynebacterium efficiens ''YS-31<br />]'''[http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?id=257309 '''Corynebacterum glutamicum'' ATCC 13032]''' | ||
|} | |} | ||
[[Image:corynebacteriumdiphteriae.jpg|frame|right|Corynebacterium diphtheriae.''From [http://www.ulb.ac.be/sciences/biodic/ImBacterie2.html BIODIC.]]] | [[Image:corynebacteriumdiphteriae.jpg|frame|right|Corynebacterium diphtheriae.''From [http://www.ulb.ac.be/sciences/biodic/ImBacterie2.html BIODIC.]]] | ||
Revision as of 18:56, 2 June 2006
|
NCBI: |

Classification
Higher order taxa:
Bacteria; Actinobacteria; Actinobacteria (class); Actinobacteridae; Actinomycetales; Corynebacterineae; Corynebacteriaceae; Corynebacterium
Species:
Corynebacterium diphtheriae; Corynebacterium efficiens; Corynebacterium glutamicum For more information on the numerous Corynebacterium species, visit NCBI.
Description and Significance
Corynebacterium diphtheriae is the etiological agent of diphtheria, an upper respiratory disease mainly affecting children. The virulence factors (most specifically diphtheria toxin) have been studied extensively and are well understood.
Genome Structure
The Corynebacterium diphtheriae NCTC 13129 genome is 2,488,625 bp in length and has an average G-C content of 53.5%. The three Corynebacterium genomes that have been sequenced are all similar in general content - visit NCBI for more information.
Cell Structure and Metabolism
Corynebacteria are small, generally nonmotile, Gram-positive, non-sporulating (although they have club-like ends), pleomorphic bacilli. Due to their snapping type of division, cells often lie in clusters resembling chinese letters. Corynebacteria are chemoorganotrophic, aerobic, or facultatively anaerobic, and they exhibit a fermentative metabolism (carbohydrates to lactic acid) under certain conditions. They are fastidious organisms, growing slowly on even an enriched medium.
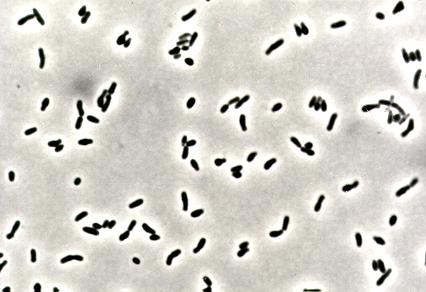
Ecology
Many species of Corynebacteria can be isolated from various places such as soil, water, blood, and human skin. Pathogenic strains of Corynebacteria can infect plants, animals, or humans. Though humans are now the only known reservoir for the disease. The bacterium is generally found in temperate zones but may also be found in other parts of the world. Nondiptherial Corynebacteria are ubiquitous in nature, and are commonly found in human mucous membranes and skin.
Isolation and Cultivation
The best method for isolating and cultivating Corynebacteria is to use sheep blood agar plus one selective medium as the primary plating media. Selective media commonly used are Cystine-Tellurite blood agar or Tinsdale medium. The plates should be ready after 18 to 24 hours of incubation at 37 degrees Celsius in a 5% carbon dioxide-enriched atmosphere.
Pathology
Diphtheria was first identified by Hippocrates in the 4th century B.C. The disease plagued Europe through the 17th, 18th, and 19th centuries. It was spread to America where it reached epidemic proportions around the middle of the 18th century. Corynebacterium diphtheriae was identified as the etiological agent of diphtheria by Klebs in 1883, and was first cultivated in 1884 by Loeffler, who also identified the diphtheria toxin in the same year.
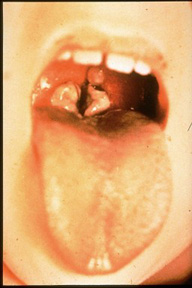
Diphtheria is described as "an upper respiratory tract illness characterized by sore throat, low-grade fever, and an adherent membrane of the tonsil(s), pharynx, and/or nose," by the CDC. The pathogenesis capabilities of diphtheria are dependent on its ability to colonize the nasopharyngeal cavity or skin and its ability to produce the diphtheria toxin. C. diphtheriae usually colonize a local lesion in the upper respiratory tract (although cutaneous diphtheria can occur as well) where the toxin secreted by the bacteria cases necrotic injury to epithelial cells. As a result, blood plasma leaks into the area and forms a fibrin network called a pseudomembrane, which is full of rapidly growing C. diphtheriae cells. At the site of the lesion the diphtheria toxin is absorbed and disseminated throughout the body via lymph channels. Most commonly affected areas include heart, muscle, peripheral nerves, adrenal glands, kidneys, liver, and spleen (rather comprehensive).
The diphtheria toxin works by causing the death of eukaryotic cells and tissues by inhibiting protein synthesis in the cells. Two key factors aid C. diphtheriae in the production of this systemic toxin: low extracellular concentrations of iron and the presence of a lysogenic prophage (talked about in detail in the phage section below). The role of iron in C. diphtheriae cultures is very dramatic, and it is assumed to play the same part in vivo as well. In iron depleted cultures C. diphtheriae will produce the diphtheria toxin as up to 5% of its total protein production. It has been found that the tox gene is regulated by a negative control. A repressor molecule, the product of the DtxR gene, is activated by iron. When activated, the repressor binds to the tox gene operator and prevents transcription.
There are three different strains of C. diphtheriae which are differentiated by the severity of the disease they cause in humans. The three strains are gravis, intermedius, and mitis (you can distinguish the severity of each strain based on its name). The difference in virulence of the three strains can be attributed to their relative abilities to manufacture the diphtheria toxin (in both rate and quantity), and their respective growth rates. The mitis strain has a generation time of about 180 minutes while the gravis strain has a generation time of about 60 minutes. This faster growth rate may allow colonies to deplete iron supplies in colonized areas faster, letting them produce toxin in greater quantities sooner.
Phages
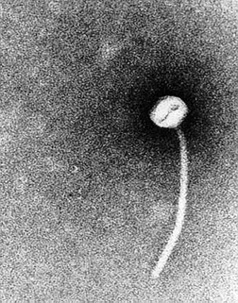
As mentioned above, a lysogenic phage plays a key role in the pathogenicity of C. diphtheriae. The tox gene, the structural gene for the diphtheria toxins carried by a family of closely related corynebacteriophages, the most extensively studied being the Beta-phage (B-phage). Interestingly, a phage lytic cycle is not required for production of the diphtheria toxin. In other words, while in most cases the phage must insert its DNA into the chromosome of the bacterium it is infecting, this is not the case with the B-phage and C. diphtheriae. The phage maintains a lysogenic life cycle, and the diphtheria toxin protein is transcribed directly from the phage DNA.
As mentioned in the pathology section, iron plays a key role in regulating the production of the toxin. The DtxR gene, however, is part of the bacterial chromosome, and toxin production is dependent on the rate of bacterial iron metabolism. So, while the bacterial chromosome does not ever actually contain the tox gene, various bacterial mechanisms still control the rate and quantity of production of the diphtheria toxin.
References
Akers, Angela S., Corynebacterium. Virginia Tech.
Frassetto, Lynda A. Corynebacterium Infections. eMedicine. November 18, 2004.
Murphy, John R. University of Texas Medical Branch: Corynebacterium Diphtheriae
Rollins, David M. University of Maryland: Pathogentic Microbiology: Corynebacterium
The Sanger Institute: Corynebacterium diphtheriae
Todar, Kenneth. University of Wisconsin-Madison Department of Bacteriology: Diphtheria
