Penicillium chrysogenum: Difference between revisions
| Line 28: | Line 28: | ||
Penicillium was originally discovered by Alexander Fleming. Fleming observed that staphylococcus cultures which had been left on the lab bench and allowed to grow, had begun to lyze and that the active factor could be extracted by filtration of the mold. He described Penicillium as a fungal colony that begins as a “white fluffy mass” that later turns green then black. A yellow color appears after several days that will diffuse throughout the medium.(2) | Penicillium was originally discovered by Alexander Fleming. Fleming observed that staphylococcus cultures which had been left on the lab bench and allowed to grow, had begun to lyze and that the active factor could be extracted by filtration of the mold. He described Penicillium as a fungal colony that begins as a “white fluffy mass” that later turns green then black. A yellow color appears after several days that will diffuse throughout the medium.(2) | ||
Penicillium chrysogenum is a common fungus that can inhabit a wide variety of habitats including the soils of degraded forests (4), on the pollen and provisions of alfalfa leafcutter bees (5), and in Arctic subglacial ice where they feed on sediment-rich basal ice shelves(6). Penicillium chrysogenum is most commonly found naturally in moist soils with plentiful quantities of carbon and nitrogen for miccohrizal growth. This species can also be found on fruit causing decay. (8) | Penicillium chrysogenum is a common fungus that can inhabit a wide variety of habitats including the soils of degraded forests (4), on the pollen and provisions of alfalfa leafcutter bees (5), and in Arctic subglacial ice where they feed on sediment-rich basal ice shelves (6). Penicillium chrysogenum is most commonly found naturally in moist soils with plentiful quantities of carbon and nitrogen for miccohrizal growth. This species can also be found on fruit causing decay. (8) | ||
The importance of sequencing the genome of Penicillium chrysogenum is evident; it is a major player in the lives of humans today in various forms; pathogen, allergen, and an industrial source of antibiotics. Therefore understanding the various metabolic and biosynthetic systems of Penicillium chrysogenum will allow researchers the ability to limit growth when it acts as a pathogen, lower the allergic response to it when it acts as an allergen, or maximize biosynthesis of penicillin when it is used to make the antibiotic. Additionally it is important to have the genome sequence of this species for analysis when considering the emergence of new drug resistant strains of bacteria. | The importance of sequencing the genome of Penicillium chrysogenum is evident; it is a major player in the lives of humans today in various forms; pathogen, allergen, and an industrial source of antibiotics. Therefore understanding the various metabolic and biosynthetic systems of Penicillium chrysogenum will allow researchers the ability to limit growth when it acts as a pathogen, lower the allergic response to it when it acts as an allergen, or maximize biosynthesis of penicillin when it is used to make the antibiotic. Additionally it is important to have the genome sequence of this species for analysis when considering the emergence of new drug resistant strains of bacteria. | ||
Revision as of 23:59, 26 August 2007
A Microbial Biorealm page on the genus Penicillium chrysogenum
Classification
Higher order taxa
Cellular organisms; Eukaryota; Fungi; Ascomycota; Eurotiomycetes; Eurotiales; Trichocomaceae; Penicillium
Species
|
NCBI: Taxonomy [1] |
|
NCBI: Genome [2] |
Penicillium chrysogenum
Description and significance
Penicillium chrysogenum is a widely studied species of Penicillium and is sometimes also known as P. notatum, P. meleagrinum,or P. cyaneofulvum (3) though occasionally they are not synonymous.(15) It plays a significant role in the medical community as an antibiotic because it can create penicillin which degrades gram positive bacteria by affecting lysis of their cell wall.(2) It can also play a role as either a pathogen (1),(9),(10), an allergen (11), and may aid in protecting crops from certain pathogenic attacks.(7)
Penicillium was originally discovered by Alexander Fleming. Fleming observed that staphylococcus cultures which had been left on the lab bench and allowed to grow, had begun to lyze and that the active factor could be extracted by filtration of the mold. He described Penicillium as a fungal colony that begins as a “white fluffy mass” that later turns green then black. A yellow color appears after several days that will diffuse throughout the medium.(2)
Penicillium chrysogenum is a common fungus that can inhabit a wide variety of habitats including the soils of degraded forests (4), on the pollen and provisions of alfalfa leafcutter bees (5), and in Arctic subglacial ice where they feed on sediment-rich basal ice shelves (6). Penicillium chrysogenum is most commonly found naturally in moist soils with plentiful quantities of carbon and nitrogen for miccohrizal growth. This species can also be found on fruit causing decay. (8)
The importance of sequencing the genome of Penicillium chrysogenum is evident; it is a major player in the lives of humans today in various forms; pathogen, allergen, and an industrial source of antibiotics. Therefore understanding the various metabolic and biosynthetic systems of Penicillium chrysogenum will allow researchers the ability to limit growth when it acts as a pathogen, lower the allergic response to it when it acts as an allergen, or maximize biosynthesis of penicillin when it is used to make the antibiotic. Additionally it is important to have the genome sequence of this species for analysis when considering the emergence of new drug resistant strains of bacteria.
Genome structure
There are four chromosomes in Penicillium chrysogenum and collectively they have a genome size of 34.1Mb.(14) The penicillin gene cluster is located on chromosome I and is 10.4 Mb in size. The genome has three genes which encode for the penicillin biosynthetic pathway, pcbAB, pcbC, and pcbDE which form a single gene cluster. This gene sequence is important to the lifestyle of this species due to the interactions it has with other microbes such as streptococcus. Without this sequence Penicillium chrysogenum would have far more competition from these microbes. The other three chromosomes are 9.6, 7.6, and 6.3 Mb in size for chromosomes II, III, and IV respectively.(15)
Many other partial sequences exist for Penicillium chrysogenum including sequences coding for sulfate uptake which requires genes sutA and sutB (16) and anthranilate synthesis by trpC.(17) However no full genome sequence exists as of Febuary 2006.(22)
Penicillium chrysogenum may also contain plasmids. Autonomously replicating plasmids that induce cotransformation have been helpful in the commercial exploitation of Penicillium chrysogenum as a penicillin maker. These plasmids can aid in over the over-expression of genes, cloning of genes, and disruption of genes.(21)
Cell structure and metabolism
Penicillium chrysogenum exhibits typical eukaryotic cell structure; a tubulin cytoskeleton which is used for motility and cell structure, a mitochondira that produces ATP via the TCA cycle, etc.(22)
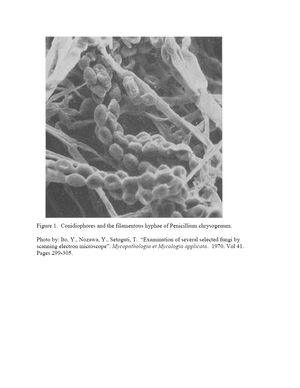
Figure 1 shows a high magnification image produced with scanning electron microscopy of the external cell structure of Penicillium chrysogenum. This image displays the typical filamentous hyphae that contain many conidia. These oblong structures are conidia, the asexual spores of the fungus.(23) These conidia are the cause of pathogenicity in human as in the cases of allergy and endophthalmitis. The conidia originate from complexes known as conidiophores. The growth of conidiophores begins when a stalk sprouts out of a foot cell. The stalk swells at the end and forms a vesicle. Sterigmata form from the vesicle which give way to long chains of conidia.(24)
An interesting aspect of the metabolism of Penicillium chrysogenum is that it will express metabolic genes differentially when grown in different medium. Preferential gene expression shuts-down secondary metabolic pathways such as the expression of Isopenicillin N synthase through PacC (P08703 gene).(22) The inactivation of PacC will also in activate the production of conidia.(24) In a glucose medium this gene is shut off while in lactose the gene is active.(22)
It has more defenses then penicillin, it has proteins that provide resistance to Fluconazole (Fluconazole resistance protein 1, P38124 gene) and cycloheximide (Cycloheximide resistance protein, P32071 gene).
Pathology
Penicillium chrysogenum is rarely pathogenic except in exceptional circumstances such as people with severely suppressed immune systems, like those with human immunodeficiency virus (HIV). Due to this low pathogenicity, it is difficult to diagnose due to a low level of suspicion of infection. Symptoms however include pulmonary infection including pneumonia, localized granulomas, fungus balls, and systemic infection. Once diagnosed, infection is treated with the surgical removal of foci of infection and the use of an oral antifungal regiment, usually either amphotericin B or itraconazole. Prognosis is poor for this type of infection. Penicillium chrysogenum is usually not a cause of infection in people with normally functioning immune systems.(1)
Another such exceptional circumstance is in the case of endophthalmitis which is described as an inflammation of the ocular cavity. The most common avenue for the implementation of infection by Penicillium chrysogenum in the eye is by penetrating trauma. The infection is treated as a systemic infection would be, with an oral antifungal regiment, usually either amphotericin B or itraconazole with the caveat that a topical antifungal may also be prescribed. (9),(10)
Penicillium chrysogenum can also act as an allergen and an asthma inducer. Pen ch 13 is the active allergen that triggers histamine responses in the epithelial cells of lungs. The constriction of the airway ensues and the characteristic hack of the asthmatic is the most common symptom. The exact mechanisms for crossing of the allergen into the epithelial cells is an active area of research.(12)
Application to Biotechnology
Penicillium chrysogenum is usually exploited for its antibiotic capabilities. It produces the hydrophobic β-lactam compound penicillin. The efficacy of the specific penicillin made is dependent on its side chain. Originally Penicillium chrysogenum was limited to the treatment of scarlet fever, pneumonia, gonorrhea, infections on wounds, and serious staphylococcal infections as Penicillian G.(13) Today many variations of side chains yield a wide variety of semi-synthetic penicillin’s that are able to fight a broader range of bacteria however Penicillium chrysogenum remains the primary producer of Penicilian G and Penicilian V.(14)
The most common application of Penicillium chrysogenum is as an industrial producer of antibiotics. However it also has recently been suggested that Penicillium chrysogenum can be used to assist crops to fight off other pathogenic species. The application of penicillin to crops such as apple trees, grapevines and tomatoes can induce their defense mechanisms and thereby help protect against apple scab, mildew, and early blite respectively in each of these crops. This response is induced under greenhouse and field conditions and is not due to direct pathogenic effects from penicillin.(7)
Current Research
The search for a complete understanding of the biosynthetic pathways for the production of penicillin in Penicillium chrysogenum is still an active area of research. It is important to understand exactly how these pathways function to maximize the industrial processes for creating penicillin. Currently there is no understanding as to how exactly iso-penicillin N (IPN) makes it into peroxisomes. It has been found that other species that secrete IPN have ABC coupled transport as in the case of Aspergillus nidulans. No such transporters exist in Penicillium chrysogenum.(18)
Quit a lot is known about the antibiotic properties of Penicillium chrysogenum however little is known regarding the antifungal properties it contains. Penicillium Antifungal Protein or PAF is a protein which inhibits the growth of certain taxonomically related filamentous fungi. PAF may also affect the permeability of the membranes of filamentous fungi by catalyzing the leaking of potassium out of the cells. Not all fungi are affected by it however and the exact action of PAF on cells is not none.(19)
Amylases have been some of the most important enzymes in the eyes of humans for thousands of years. They are a required work horse in the processes of alcoholic fermentation. Because of their importance, they have been subject to industrial production. Therefore new cost effective ways for producing amylases are studied. One way to produce α-amylase (one of two types of amylases) is by Solid State Fermentation (SSF) which is a process that exposes an insoluble substrate to moisture but not standing water for fermentation. Cheap agricultural by-products such as wheat bran and sunflower oil meal in combination have been fermented with Penicillium chrysogenum which produces α-amylase. This is a low cost way to produce the product however large scale experiments are needed to ensure industrial application.(20)
References
1. Adrian, B.L., Burdette, S.D., and Herchline, T.E. "Intestinal invasion and disseminated disease associated with Penicillium chrysogenum". Ann Clin Microbiol Antimicrob. December 2005. Vol 4. Page 21.
http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=1343575
2. Fleming, A. "On the antibacterial action of cultures of Penicillium, with special refrence to their use in the isolation of B. Influenzae". British Journal of Experimental Pathology. May 1929. Vol 10. Pages 226-236.
3. Samson, R.A., Hadlok, R.,and Stolk, A.C. "A taxonomic study of the Penicillium chrysogenum series". Antonie van Leeuwenhoek. May 1977. Vol 43. Pages 169-175
http://www.springerlink.com/content/wu67kx6758061h87/fulltext.pdf
4. Jka, D.K., Sharma, G.D., and Mishra R.R. "Ecology of soil microflora and mycorrihzal symbionts in degraded forests at two altitudes". Biology and Fertility of Soils. January 1992. Vol 12. Pages 272-278.
http://www.springerlink.com/content/u85m3v54g29w85u2/fulltext.pdf
5. Inglis, G.D., Sigler, L., and Goette, M.S. "Aerobic microorganisims associated with alfalfa leafcutter bees (Megachile rotundata)". Microbial Ecology. September 1993. Vol 26. Pages 125-143.
http://www.springerlink.com/content/q73337l00677x668/fulltext.pdf
6. Sonjak, S., Frisvad, J.C., and Gunde-Cimerman, N. "Penicillium mycobiota in Arctic subglacial ice". Microbial Ecology. August 2006. Vol 52. Pages 207-216.
http://www.springerlink.com/content/n8m5266050067r11/fulltext.pdf
7. Thuerig, B., Binder, A., Boller, T., et al. "An aqueous extract of the dry mycelium of Penicillium chrysogenum induces resistance in several crops under controlled and field conditions". European Journal of Plant Pathology. Febuary 2006. Vol 114. Pages 185-197.
http://springerlink.metapress.com/content/x6x28h52507857m1/fulltext.pdf
8. Barkai-Golan, R. "Species of Penicillium causing decay of stored fruit in Isreal". Mycopathologia. October 1974. Vol 54. Pages 141-145.
http://www.springerlink.com/content/x530n77737132218/fulltext.pdf
9. Eschete, M.L., King, J.W., West, B.C., et al. "Penicillium chrysogenum endophthalmitis". Mycopathologia. May 1981. Vol 74. Pages 125-127.
http://www.springerlink.com/content/x1311078w8k21343/fulltext.pdf
10. Galland, F., Le Goff, L., Conrath, J., et al. "Penicillium chrysogenum endophthalmitis: a case report". Journal Français d'Ophtalmologie. March 2004. Vol 27. Pages 264-266.
11. Shen, H.D., Chou, H., Tam, M.F., et al. “Molecular and immunological characterization of Pen ch 18, the vacuolar serine protease major allergen of Penicillium chrysogenum” Allergy. 2003 Oct;58(10):993-1002
http://www.ncbi.nlm.nih.gov/sites/entrez?db=PubMed&cmd=Retrieve&list_uids=14510716
12. H.-Y. Tai, M. F. Tam, H. Chou, H.-J. Peng, S.-N. Su, D.-W. Perng, H.-D. Shen. “Pen ch 13 allergen induces secretion of mediators and degradation of occludin protein of human lung epithelial cells”. Allergy. March 2006. Vol 61. Pages 382–388.
13. Demain, A.L., Elander, R.P. “The β-lactam antibiotics: past, present, and future”. Antonie Van Leeuwenhoek. September 1998. Vol 75. Pages 5-19.
http://springerlink.metapress.com/content/ukn8g13608710123/fulltext.pdf
14. Xu, Z., A. van den Berg, M., Scheuring, C., et al. “Genome physical mapping from large-insert clones by fingerprint analysis with capillary electrophoresis: a robust physical map of Penicillium chrysogenum”. Nucleic Acids Research. March 2005. Vol 33. No 5. http://nar.oxfordjournals.org/cgi/screenpdf/33/5/e50
15. Fierro, F., Gutierrez, S., Diez, B., et al. “Resolution of four large chromosomes in penicillin-producing filamentous fungi: the penicillin gene cluster is located on chromosome II (9.6 Mb) in Penicillium notatum and chromosome I 10.4 Mb) in Penicillium chrysogenum”. MGG. January 1993. Vol 241. Pages 573-578
http://springerlink.metapress.com/content/hp01726x273w0577/fulltext.pdf
16. Mart van de Kamp, Pizzinini, E., Vos, A., et al. “Sulfate Transport in Penicillium chrysogenum: Cloning and Characterization of the sutA and sutB Genes”. Journal of Bacteriology. September 1999. Vol 23. Pages 7228-7234.
http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=103684
17. Sanchez, F., Tourino, A., Traseira, S., et al. “Molecular cloning and characterization of the trpC gene from Penicillium chrysogenum”. MGG. 1986. Vol 205. Pages 248-252.
http://springerlink.metapress.com/content/vu836185hv701211/fulltext.pdf
18. Garcia-Estrada, C., Vaca, I., Lamas-Maceriras, M., et al. “In vivo transport of the intermediates of the penicillin biosynthetic pathway in tailored strains of Penicillium chrysogenum”. Applied Genetics and Molecular Biotechnology. May 2007.
http://springerlink.metapress.com/content/65280m822km0n846/fulltext.html
19. Kaiserer, L., Oberparleiter, C., Weiler-Gorz, R., et al. “Charecterization of the Penicillium chrysogenum antifungal protein PAF”. Archives of Microbiology. September 2003. Vol 180. Pages 204-210.
http://springerlink.metapress.com/content/b1anupgmv3hm4w7f/fulltext.html
20. Ertan, F., Balkan, B., Balkan, S., et al. “Solid state fermentation for the production of α-amylase from Penicillium chrysogenum using mixed adricultural by-products as substrate”. Biologia. December 2006. Vol 61. Pages 657-661
http://springerlink.metapress.com/content/n38l520546786176/fulltext.pdf
21. Bañuelos, O., Naranjo, L., Casqueiro, J., et al. “Co-transformation with autonomous replicating and integrative plasmids in Penicillium chrysogenum is highly efficient and leads in some cases to rescue of the intact integrative plasmid”. Fungal Genetics and Biology. November 2003. Pages 83-92.
22. Castillo, N.I.,Fierro, F., Gutiérrez, S., et al. “Genome-wide analysis of differentially expressed genes from Penicillium chrysogenum grown with a repressing or a non-repressing carbon source”. Current Genetics. Febuary 2006. Vol 49. Pages 85-96.
http://www.springerlink.com/content/f1394783l02p7045/fulltext.pdf
23. Ito, Y., Nozawa, Y., Setoguti, T. “Examination of several selected fungi by scanning electron microscope”. Mycopathologia et Mycologia applicata. 1970. Vol 41. Pages 299-305.
http://www.springerlink.com/content/ww0x48q4763765u7/fulltext.pdf
24. Calvo, A.M., Wilson, R.A., Bok, J.W. “Relationship between secondary metabolism and fungal development”. September 2002. Vol 66. Pages 447-459.
http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=120793