Xanthomonas campestris: Difference between revisions
| Line 4: | Line 4: | ||
==Classification== | ==Classification== | ||
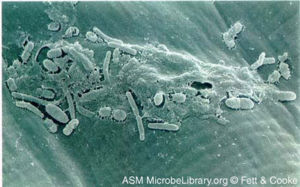
[[Image:Xanthomonas_campestris_biofilm.jpg|right|300px|thumb|''Xanthomonas campestris' biofilm on plant surface''<br> | |||
Courtesy of Fett & Cooke, ASM MicrobeLibrary [http://www.microbelibrary.org/asmonly/details.asp?id=501&Lang=]]] | Courtesy of Fett & Cooke, ASM MicrobeLibrary [http://www.microbelibrary.org/asmonly/details.asp?id=501&Lang=]]] | ||
===Higher order taxa=== | ===Higher order taxa=== | ||
Revision as of 19:33, 27 August 2007
A Microbial Biorealm page on the genus Xanthomonas campestris
this page is still under construction.
Classification
Higher order taxa
[Kingdom] Bacteria
[Phylum] Proteobacteria
[Class] Gamma Proteobacteria
[Order] Xanthomonadales
[Family] Xanthomonadaceae
[Genus] Xanthomonas
[Species] Xanthomonas campestris
Species
|
NCBI: Taxonomy |
Xanthomonas campestris
Description and significance
Xanthomonas campestris is an aerobic, Gram-negative rod known to cause the black rot in crucifers by darkening the vascular tissues. Host associated, over 20 different pathovars of X. campestris have been identified by their distinctive pathogenicity on a wide range of plants including crops and wild plants. This bacterium is mesophilic with optimal temperature at 25-30 degrees Celsius (77-85 degrees Fahrenheit). It is inactive at temperatures below 10 degrees Celsius (50 degrees Fahrenheit). [1] X. campestris have long pilus that helps them glide through water. They can live in a soil for over a year and spread through irrigation and surface water. Spraying healthy plants with copper fungicides may reduce the spread of the bacteria in the field. However, once the plant has been infected, X. campestris will eventually spread to the seed stalk inhibiting the growth of a healthy offspring.
By pure culture fermentation, X. campestris can produce an extracellular polysaccharide known as xanthan gum that is commercially manufactured as a stabilizing agent used in many industries. This organism is a model organism for plant pathogens because of their interaction between hosts. Due to the deficit in crops, further study of X. campestris’ genome may provide a solution to make plants resistant to this pathogen.
Genome structure
X. campestris have circular chromosomes containing at least two plasmids. The genome structures of X. campestris contain variation depending on the pathovars. However, the different strains of X. campestris exhibit similar characteristics like the mobile genetic elements and protein coding sequences. Over 4500 genes have been found in the genome to encode proteins however over one-third has no known function. In X. campestris pv. campestris (Xcc) wild-type strain B100, it is found to contain a plasmid which contains 3-4 kb of chromosomal fragments. [?? (2)] With over 548 unique coding sequences, X. campestris pv. Vesicatoria (Xcv) is composed of a 5.17-Mb circular chromosome, four plasmids, and an essential type III protein secretion system for pathogenicity. (3) Using Recominbase-based In Vivo Expression Technology to target tomato, Xcv has been found to have 61 genes that are involved in the interaction between pathogen and host including a necessary virulence transporter, citH homologue gene (4).
Cell structure and metabolism
X. campestris is an aerobic, rod-shaped Gram-negative bacteria characterized by its two cell walls and yellow pigment. It has a filamentous structure of pili that provides motility in water, appendage to cell surface, and also a way to transfer bacterial proteins to the plant. Proteins found in X. campestris include XpsD protein which is required for extracellular protein secretion. X. campestris contains a type III secretion system (TTSS) that is necessary in order for this pathogen to attack the host. [?? (5)] TTSS is important in pathogenesis because it delivers effector proteins into the host cell. These effectors may behave avirulently by disguising itself to secrete several hypersensitive reaction and outer proteins in order for interaction to occur with the host cells. (6) X. campestris is known to use cell-cell communication through diffusible signal factor (DSF). (7)
Ecology
Describe any interactions with other organisms (included eukaryotes), contributions to the environment, effect on environment, etc.
Pathology
How does this organism cause disease? Human, animal, plant hosts? Virulence factors, as well as patient symptoms.
Application to Biotechnology
X. campestris is used to create a stabilizing agent called xanthan gum. It was first produced at Kelco Company, a major pharmaceutical company. This preservative is an ingredient in products like Kraft French dressing, Weight Watchers food, Wonder Bread products, and more. (11) Fermented from glucose by X. campestris, xanthan gum is used to extend freshness for food products. In addition, xanthan gum also prolongs oil and gas wells even after production. Either pumped into the ground or using high pressure sandblasting, mixing water and xanthan gum into the wells will help release crude products of oil and cut through rocks in gas and oil wells. Xanthan gum costs $7 per pound compared to cornstarch for 89 cents per pound. (11)
Current Research
Genome sequence is being done in search of the essential genes needed in order to develop resistant plants.
Current research is being done on the genetic diversity in Xcc of wild crucifers. With the most diverse and abundant wild cruciferous plants in the world, research was done in California to find any differences in genetic strains on Xcc in infected wild weeds. From both non-cultivated and cultivated areas, Xcc was isolated from different regions of California. Using Amplified fragment length polymorphism PCR (AFLP) to identify genetic variation in strains, over 72 strains were sequenced to show 7 unique genotypes that were limited to their respective sites. Non-cultivated wild weeds near the coast had strains of Xcc that were specific to the region and different from the weeds grown near produced crop areas. [?? (12)]
Recent students have shown X. campestris use the diffusible signal factor (DSF) for cell-cell signaling. In order for microcolonies to form structured biofilm, synthesis of DSF require genes from the regulation of pathogenicity factor (Rpf) cluster. Without the critical DSF signalling, X. campestris will create an unstructured biofilm of bacteria. (13)
References
1.) Averre, Charles W. "Black Rot of Cabbage and Related Crops" http://www.ces.ncsu.edu/depts/pp/notes/oldnotes/vg16.htm
2.) Ko-Hsin Chin, Wei-Tien Kuo, Chia-Cheng Chou, Hui-Lin Shr, Ping-Chiang Lyu, Andrew H.-J. Wang, Shan-Ho Chou “Cloning, purification, crystallization and preliminary X-ray analysis of XC229, a conserved hypothetical protein from Xanthomonas campestris” Acta Crystallographica Section F 61 (7), 694–696.
3.) Thieme F, Koebnik R, Bekel T, et al. "Insights into genome plasticity and pathogenicity of the plant pathogenic bacterium Xanthomonas campestris pv. vesicatoria revleaed by the complete genome sequence." Bacteriol. 2005 Nov. p 7254-66. http://www.ncbi.nlm.nih.gov/sites/entrez?cmd=Retrieve&db=pubmed&list_uids=16237009
4.) Tamir Ariel D., Navon N, Burdman S. "Identification of Xanthomonas campestris pv. vesicatoria Genes Induced in its interaction with tomato." J Bacteriol. 2007 Jun 15
http://www.ncbi.nlm.nih.gov/sites/entrez?Db=pubmed&Cmd=ShowDetailView&TermToSearch=17573477&ordinalpos=11&itool=EntrezSystem2.PEntrez.Pubmed.Pubmed_ResultsPanel.Pubmed_RVDocSum
5.) ??
6.) Wang L, Tang X, He C. "The bifunctional effector AvrXccC of Xanthomonas campestris pv. campestris requires plasma membrane-anchoring for host recognition." Molecular Plant Pathology. 2007 July. p 491–501. http://www.blackwell-synergy.com/doi/abs/10.1111/j.1364-3703.2007.00409.x?prevSearch=allfield%3A%28xanthomonas+campestris%29
8.) Ritchie David F, Averre Charles W. "Bacterial Spot of Pepper and Tomato." North Carolina State University College of Agriculture and Life Sciences. 1996 June. http://www.ces.ncsu.edu/depts/pp/notes/Vegetable/vdin018/vdin018.htm
9.) Niehaus Karsten. "The Xanthomonas campestris pv. campestris genome project." https://www.genetik.uni-bielefeld.de/GenoMik/partner/bi_niehaus.html
10.) da Silva AC, Ferro JA, Reinach FC, et al. "Comparison of the genomes of two Xanthomonas pathogens with differing host specificities". Nature. 2002 May 23. p. 459-63 http://www.ncbi.nlm.nih.gov/sites/entrez?cmd=Retrieve&db=PubMed&list_uids=12024217&dopt=AbstractPlus
11.) http://www.ars.usda.gov/business/docs.htm?docid=769&page=5
12.) Ignatov, A., Sechler, A.J., Schuenzel, E., Agarkova, I.V., Vidaver, A.K., Oliver, B., Schaad, N.W. 2007. "Genetic diversity in populations of Xanthomonas campestris pv. camestris in cruciferous weeds in central coastal California". Phytopathology. 97:803-812
13.) Torres PS, Malamud F, Rigano LA, Russo DM, et al. "Controlled synthesis of the DSF cell-cell signal is required for biofilm formation and virulence in Xanthomonas campestris." Environ microbiol. 2007 Aug. p 2101-9. http://www.ncbi.nlm.nih.gov/sites/entrez?Db=pubmed&Cmd=ShowDetailView&TermToSearch=17635553&ordinalpos=1&itool=EntrezSystem2.PEntrez.Pubmed.Pubmed_ResultsPanel.Pubmed_RVDocSum
Edited by Tammie Chau, student of Rachel Larsen