Ralstonia solanacearum: Difference between revisions
| Line 33: | Line 33: | ||
==Cell structure and metabolism== | ==Cell structure and metabolism== | ||
<i>Ralstonia solanacearum</i> is a Gram negative bacteria. It has an outer membrane and inner membrane. Its cell walls consist of peptidoglycan (6). The outer membrane allows more resistance to toxins that could damage the pathogen. Motility is accomplished by pili and flagella, allowing the invasion and colonization of the pathogen into their host. Once inside the host, they remain immobile (10). | <i>Ralstonia solanacearum</i> is a Gram negative bacteria. It has an outer membrane and inner membrane. Its cell walls consist of peptidoglycan (6). The outer membrane allows more resistance to toxins that could damage the pathogen. Motility is accomplished by pili and flagella, allowing the invasion and colonization of the pathogen into their host. Once inside the host, they remain immobile (10). | ||
Revision as of 19:31, 28 August 2007
A Microbial Biorealm page on the genus Ralstonia solanacearum
Classification
Higher order taxa
Bacteria; Proteobacteria; Beta Proteobacteria; Burkholderiales; Ralstoniaceae
Species
Ralstonia solanacearum
Description and significance
caption: Wilting of a plant leaf as a result of the invasion of R. solanacearum (3).
Ralstonia solanacearum is a plant pathogenic bacterium. This organism is responsible for bacterial wilts and infection of over 200 plants species, and its effect can be observed by the appearance of wilting of plants (1). It is usually found in soils of tropical and subtropical countries. This pathogen can lie dormant in water or soil until a host plant grows. Once the host begins to develop, the organism spreads throughout the plant by entering the roots and colonizing water-conducting vessels (1).
Through genome sequencing, we observe that the genome encodes many proteins potentially associated with a role in pathogenicity. In particular, many putative attachment factors were identified (2). The analysis of the genome has made it possible to identify more than 200 new genes that are potentially involved in virulence (4).
Genome structure
R. solanacearum genome is segregated into two circular molecules: a large replicon of 3,716,413 bp and a smaller 2,094,509-bp replicon, yielding a total genome size of 5,810,922 bp (5). The two molecules have an almost identical G+C content (67.04% and 66.86% for the large and small replicon, respectively)(5). The genome encodes many proteins potentially associated with a role in pathogenicity.
The complete genome sequence and its analysis of R. solanacearum strain GMI1000 shows that it possesses the 5.8-megabase (Mb) genome, which is organized into two replicons: a 3.7-Mb chromosome and a 2.1-Mb megaplasmid (5).
The genes that are present on the megaplasmid suggests that this replicon has a significant function in overall fitness and adaptation of the bacterium to various environmental conditions (9). This validates that the megaplasmid carry all the information necessary for plant pathogenesis.
Cell structure and metabolism
Ralstonia solanacearum is a Gram negative bacteria. It has an outer membrane and inner membrane. Its cell walls consist of peptidoglycan (6). The outer membrane allows more resistance to toxins that could damage the pathogen. Motility is accomplished by pili and flagella, allowing the invasion and colonization of the pathogen into their host. Once inside the host, they remain immobile (10).
The plant pathogen Ralstonia solanacearum needs aerotaxis for normal biofilm formation. In order to locate and infect host plant roots, R. solanacearum needs taxis, the ability to move towards more favorable conditions. Aerotaxis, or energy taxis, which guides bacteria toward optimal intracellular energy levels. The energy taxis contributes significantly to the ability of R. solanacearum to locate and effectively interact with its host plants (7).
It is known to produce a potent L-fuctose-binding lectin, RSL, a small protein of 90 amino acids with a tandem repeat in its amino acid sequence. The RSL forms a trimer which produces a six-bladed Beta-propellor (6). Host plant cell walls contain carbohydrates which enable attachment sites for the pathogen protein receptors. In this case, the lectin has a great affinity for αFuc1-2Gal and αFuc1-6Gal epitopes found on certain plants (6).
Ecology
Ralstonia solanacearum is commonly found in the soils of tropical and subtropical countries where it devastates cultures of many crop plants. Certain strains are adapted to milder environmental conditions and have recently been isolated in northern European countries and also found in warm temperate regions. Major agricultural hosts include tobacco, tomato, potato, eggplant, pepper and banana trees (1). This bacterium can also be free-living as a saprophyte in water or in the soil in the absence of host plants (8).
R. solanacearum has the ability to survive for long periods of time in a nutrient-depleted bulk soil environment. Due to its very broad host range of wide geographical distribution, it is arguably the world's single most harmful bacterial plant pathogen. Bacterial wilt has a disproportionately high impact on subsistence and small-scale growers in the developing world (11).
Pathology
R. solanacearum possesses hrp encoding the type III secretion system (T3SS), and its pathogenicity depends on interactions between the host plant and type III effectors (9). R. solanacearum invades intercellular spaces of roots through openings such as wounds then accumulates around the stele before breaking into and filling the xylem vessels. On invasion of the xylem vessels, the bacteria grow and travel rapidly to the upper parts of the plant. This results in extensive wilting because of reduced sap flow caused by the presence of a large number of bacteria cells and exopolysaccharide (EPS) slime produced by the bacteria in some xylem vessels (9). The main virulence factor of R. solanacearum is therefore thought to be EPS(9).
R. solanacearum also produces several known virulence factors including a consortium of plant cell wall-degrading enzymes (CWDEs), which are secreted via the type II secretion system (T2SS) (9). The symptoms of plants that are infected with this pathogen is evident by observed physical wilting.
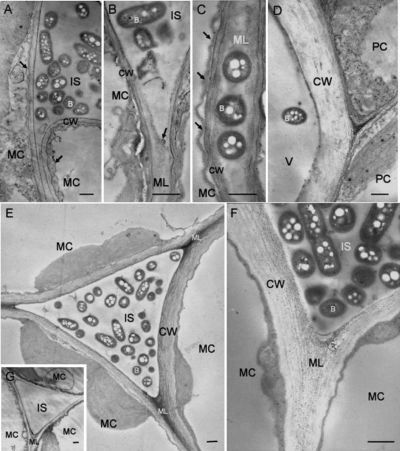
caption: Electron micrographs showing tobacco leaf tissues at 3 days after infiltration with R. solanacearum (9).
Application to Biotechnology
The proliferation of the Ralstonia solanacearum in intercellular spaces is the quantitative determinant of its pathogenicity and is dependent on hrp genes. R. solanacearum secretes PopA, an extracellular Hrp protein and harpin, which contain high proportions of glycine and alanine. After proliferation in intercellular spaces, the bacteria systemically infect the host plants through xylem vessels and produces EPS, which is involved in quantitatively controlling virulence (9).
Ralstonia solanacearum also produces a potent L-fuctose-binding lectin, RSL, a small protein of 90 amino acids with a tandem repeat in its amino acid sequence. As a result, the lectin protein has an high affinity for the αFuc1-2Gal containing oligosaccharides on certain plants (6).
Current Research
1) A current study was performed on Ralstonia solanacearum biovar 2 and its survival dependent on a variation of temperatures. The pathogen was recovered at low densities (10 to 80 CFU/ml) by direct plating on modified SMSA agar from water samples at 14°C or higher, but its isolation was usually unsuccessful at temperatures below 9°C (12). To study them in the winter, two liquid selective media for enrichment (at 29 and 35°C) were utilized and compared them by using spiked river water samples; Detection of R. solanacearum was carried out by a double-antibody-sandwich indirect enzyme-lined immunosorbent assay (DASI-ELISA) with modified Wilbrink broth (MWB) (12). The best detection results were obtained by the most-probable-number method after enrichment at 35°C with MWB (12). Overall, R. solanacearum is able to survive at low temperatures.
2) A recent study of Ralstonia solanacearum utilizing qualitative and quantitative chemotaxis assays revealed that this bacterium is specifically attracted to diverse amino acids and organic acids (13). R. solanacearum pathogens that lack flagellas and do not possess motility due to mutation is significantly reduced in virulence (13). Taxis makes its contribution to virulence in the early stages of host invasion and colonization because nontactic strains were as virulent as the wild-type strain was when bacteria were introduced directly into the plant stem (13). When inoculated individually by soaking the soil, both nontactic mutants reached the same population sizes as the wild type did in the stems of tomato plants just beginning to wilt. However, when tomato plants were coinoculated with a 1:1 mixture of a nontactic mutant and its wild-type parent, the wild-type strain outcompeted both nontactic mutants by 100-fold (13). These observations show that chemotaxis is an essential requirement for virulence and competitive, pathogenic fitness in Ralstonia solanacearum.
3) A current research based on Ralstonia solanacearum encoding a family of seven type III secretion system (T3SS) effectors that contain both a leucine-rich repeat and an F-box domain (14). A R. solanacearum strain in which all of the seven GALA effector genes have been deleted or mutated was no longer pathogenic (14). GALA T3SS effectors are essential to R. solanacearum to control disease (14). The F-box domain is an important contribution to the virulence function of GALA7. Therefore, these effectors act by hijacking their host SCF-type E3 ubiquitin ligases to interfere with their host ubiquitin/proteasome pathway to promote disease (14).
References
Edited by Cam-Tu Dang, student of Rachel Larsen
1. Vailleau, F., Sartorel, E., Jardinaud, M.F., Chardon, F. Genin, S., Huguet, T., Gentzbittel, L., Petitprez, M. (2007). "Characterization of the interaction between the bacterial wilt pathogen Ralstonia solanacearum and the model legume plant Medicago truncatula." Molecular Plant-Microbe Interactions 20: 159-167.
2. Salanoubat M, Genin S, Artiguenave F, Gouzy J, Mangenot S, Arlat M, Billault A, Brottier P, Camus JC, Cattolico L, Chandler M, Choisne N, Claudel-Renard C, Cunnac S, Demange N, Gaspin C, Lavie M, Moisan A, Robert C, Saurin W, Schiex T, Siguier P, Thébault P, Whalen M, Wincker P, Levy M, Weissenbach J, Boucher CA. "Genome sequence of the plant pathogen Ralstonia solanacearum". PubMed. 2002. p. 497-502. <http://www.ncbi.nlm.nih.gov/sites/entrez?cmd=Retrieve&db=PubMed&list_uids=11823852&dopt=AbstractPlus&holding=f1000%2Cf1000m%2Cisrctn>
3. Williams-Woodward, J. "Southern Bacterial Wilt". Invasive Species. 2003. <http://www.invasive.org/browse/detail.cfm?imgnum=9000025>.
4. Boucher, C. "Complete genome sequence of the plant pathogen bacterium Ralstonia solanacearum sheds light on the mechanisms governing pathogenicity". Nature. 2002. <http://www.cnrs.fr/cw/en/pres/compress/Ralstonia.htm>
5. M. Salanoubat, S., Genin, F., Artiguenave, J., Gouzy, S., Mangenot, M., Arlat, A., Billault, P., Brottier, J. C., Camus, L., Cattolico, M., Chandler, N., Choisne, C., Claudel-Renard, S., Cunnac, N., Demange, C., Gaspin, M., Lavie, A., Moisan, C., Robert, W., Saurin, T., Schiex, P., Siguier, P., Thébault, M., Whalen, P., Wincker, M., Levy, J., Weissenbach and C., A. Boucher. "Genome sequence of the plant pathogen Ralstonia solanacearum". International Weekly Journal of Science. 2002. <http://www.nature.com/nature/journal/v415/n6871/full/415497a.html>
6. Kostlánová1, N., Mitchell, E., Lortat-Jacob, H., Oscarson, S., Lahmann, O., Gilboa-Garber, N., Chambat, G., Wimmerová, M., Imberty, A. "THE FUCOSE-BINDING LECTIN FROM RALSTONIA SOLANACEARUM: A NEW TYPE OF β-PROPELLER ARCHITECTURE FORMED BY OLIGOMERISATION AND INTERACTING WITH FUCOSIDE, FUCOSYLLACTOSE AND PLANT XYLOGLUCAN". JBC Papers. 2005. <http://www.jbc.org/cgi/reprint/M505184200v1.pdf>
7. Yao, J., Allen, C. "The plant pathogen Ralstonia solanacearum needs aerotaxis for normal biofilm formation and interactions with its tomato host". Journal of Bacteriology. 2007. <http://jb.asm.org/cgi/content/abstract/JB.00398-07v1>
8. Genin, S., and Boucher, C. "LESSONS LEARNED FROM THE GENOME ANALYSIS OF RALSTONIA SOLANACEARUM". Annual Review of Phytopathology. Vol. 42: 107-134. 2004. <http://arjournals.annualreviews.org/doi/abs/10.1146/annurev.phyto.42.011204.104301>
9. Hikichi1,Y., Yoshimochi, T., Tsujimoto, S., Shinohara, R., Nakaho, K., Kanda, A., Kiba, A., Ohnishi, K. "Global regulation of pathogenicity mechanism of Ralstonia solanacearum". Plant Biotechnology. p. 149–154. 2007. <http://www.jspcmb.jp/pbcontents/pdf/pb24_1/24_149.pdf>
10. Tans-Kersten, J., Brown, D., Allen, C. "Swimming Motility, a Virulence Trait of Ralstonia solanacearum, is Regulated by FIhDC and the Plant Host Environment". The American Phytopathological Society. Vol 17. p. 686-695. 2004. <http://www.plantpath.wisc.edu/fac/cza/Tans-Kersten%20et%20al%20'04.pdf>
11. Allen, C. "Genetics of bacterial wilt virulence". Plant Pathology and Women's Studies. 2007. <http://www.plantpath.wisc.edu/fac/cza.htm>
12. Caruso, P., Palomo, J., Bertolini, E., Álvarez, B., López, M., Biosca, E. "Seasonal Variation of Ralstonia solanacearum Biovar 2 Populations in a Spanish River: Recovery of Stressed Cells at Low Temperatures". American Society for Microbiology. 2005. Vol. 71. p. 140-148. <http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=544205&rendertype=abstract>
13. Yao, J., Allen, C. "Chemotaxis Is Required for Virulence and Competitive Fitness of the Bacterial Wilt Pathogen Ralstonia solanacearum". American Society for Microbiology. 2006. Vol 188. p. 3697–3708. <http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=1482862&rendertype=abstract>
14. Angot, A., Peeters, N., Lechner, E., Vailleau, F., Baud, C., Gentzbittel, L., Sartorel, E., Genschik, P., Boucher, C., Genin, S. "Ralstonia solanacearum requires F-box-like domain-containing type III effectors to promote disease on several host plants". The National Academy of Sciences of the USA. 2006. Vol. 103. p. 14620–14625. <http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=1600009>