Pneumocystis carinii: Difference between revisions
| Line 34: | Line 34: | ||
==Cell structure and metabolism== | ==Cell structure and metabolism== | ||
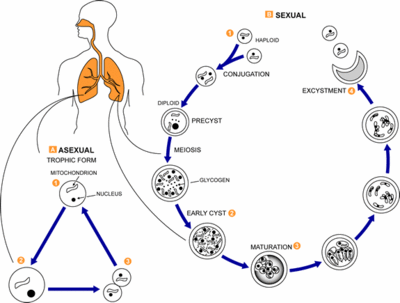
[[Image:Pneumocystis_LifeCycle.gif|thumb|Pneumocystis Life Cycle|400px|right|Life cycle of ''Pneumocystis carinii''. From [http://www.dpd.cdc.gov/dpdx/HTML/Pneumocystis.htm Centers for Disease Control and Prevention Division of Parasitic Diseases (DPDx).]]] | [[Image:Pneumocystis_LifeCycle.gif|thumb|Pneumocystis Life Cycle|400px|right|Life cycle of ''Pneumocystis carinii''. From [http://www.dpd.cdc.gov/dpdx/HTML/Pneumocystis.htm Centers for Disease Control and Prevention Division of Parasitic Diseases (DPDx).]]] | ||
The metabolism of lipids by ''Pneumocystis carinii'' has been unlocked by the use of radiolabeled precursors in purified samples of the organism. ''P. carinii'' is capable of incorporating ethanolamine and choline precursors into phospholipid head groups. Additionally, it was found that the organism is capable of metabolizing PE to PC by the methylation of the glycerophospholipid head group. Experiments using radiolabeled serine and inositol have established that ''P. carinii'' might be able to convert PS to PE. | |||
==Ecology== | ==Ecology== | ||
Revision as of 04:55, 29 August 2007
A Microbial Biorealm page on the genus Pneumocystis carinii
Classification
Higher order taxa
Eukaryota; Fungi; Dikarya; Ascomycota; Taphrinomycotina; Pneumocystidomycetes; Pneumocystidaceae; Pneumocystis
Species
Pneumocystis carinii
|
NCBI: Taxonomy |
Description and significance
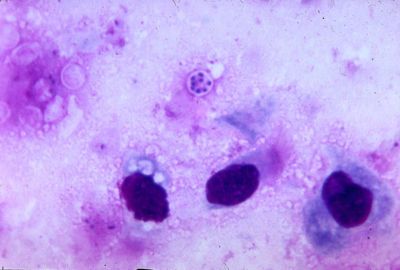
From the genus Pneumocystis, Pneumocystis carinii, is a fungal pathogen which causes the disease Pneumocystis pneumonia (PCP) in immunocompromised mammalian hosts, specifically rats. Pneumocystis carinii was originally thought to be a protozoon when it was discovered in 1912 at the Pasteur Institute. However, in 1988, the organism was reclassified as a member of the fungal kingdom after its small ribosomal RNA subunit had been sequenced. The full genome has yet to be sequenced, but progress is currently underway, see Genome structure (1). The initial classification of Pneumocystis carinii as a protozoa was partly due to the life-cycle of the organism.
Although Pneumocystis carinii has yet to be grown in pure culture, aspects of its life-cycle have been observed within infected mammalian lung tissue (see cell structure). During its known life-cycle, P. carinii can be found as trophic and cystic forms (2).
The HIV pandemic elevated interest in Pneumocystis carinii since it is the pathogen which caused Pneumocystis pneumonia and consequently a major cause of mortality in patients diagnosed with AIDS (see pathology). Despite successes against P. carinii with Trimethoprim with sulfamethoxazole there are signs that drug target sights may be mutating (1).
It should be noted that both Pneumocystis carinii and Pneumocystis jirovecii currently refer to the same organism in medical literature with regards to PCP. P. jirovecii is the organism isolated from humans while P. carinii is found in rats(3). The two organisms cause PCP in their respective hosts.
Genome structure
Sequencing of the Pneumocystis carinii is currently in progress and knowledge about the gnome has been advanced by the Pneumocystis Genome Project and the Wellcome Trust Sanger Institute Pneumocystis carnii telomeric clone sequencing project. Information from these two programs has not been made directly available to the public at this point in time.
The P. carinii genome is contained by 15 chromosomes which are from 300 kb to 700 kb in size. On these 15 chromosomes there are approximately 8 million base pairs of DNA. One feature of the P. carinii genome is that the organism has a high concentration of adenine and thymine bases (65% of the total base pairs). The genome has introns contain 38 to 424 base pairs and there have been up to nine introns per gene. The yeast Schizosaccharomyces pombe has been used as a homologue for the expressed sequenced tags (ESTs) found within the P. carinii genome (4). Completion of genomic sequence will prove invaluable in unlocking the biology of Pneumocystis carinii beyond what has been determined through the study of infected mammalian lung tissue.
Cell structure and metabolism
The metabolism of lipids by Pneumocystis carinii has been unlocked by the use of radiolabeled precursors in purified samples of the organism. P. carinii is capable of incorporating ethanolamine and choline precursors into phospholipid head groups. Additionally, it was found that the organism is capable of metabolizing PE to PC by the methylation of the glycerophospholipid head group. Experiments using radiolabeled serine and inositol have established that P. carinii might be able to convert PS to PE.
Ecology
P. carinii is found in the lung tissue of immunocompetent and immunocompromised hosts. The organism adheres specifically to the epithelium of the alveoli within the lug. Only hosts with compromised immune systems are at risk of developing PCP from P. carinii while a normal host will mount an adequate immune response to contain P. carinii.
Fragments of DNA identical to DNA sequences found in P. carinii have been found in ambient air(5) and pond water (6). Even though P. carinii nucleic acids have been found outside of the host environment, there is no definitive evidence to support an ecological niche for Pneumocystis carinii outside of a host environment.
Pathology
How does this organism cause disease? Human, animal, plant hosts? Virulence factors, as well as patient symptoms.
Application to Biotechnology
Does this organism produce any useful compounds or enzymes? What are they and how are they used?
Current Research
Enter summaries of the most recent research here--at least three required
References
Edited by Daniel Spisak of Rachel Larsen
