Mitochondria: Difference between revisions
No edit summary |
No edit summary |
||
| Line 1: | Line 1: | ||
{| | {| | ||
| width="97" height="69" bgcolor="#FFDF95" align="center" | | | width="97" height="69" bgcolor="#FFDF95" align="center" | | ||
'''NCBI:<br />[http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Info&id=780&lvl=3&lin=f&keep=1&srchmode=1&unlock Taxonomy]<br /> Genome<br /> (examples):<br /> -''<font size="2">[http://www.ncbi.nlm.nih.gov/genomes/framik.cgi?db=genome&gi=17601 Drosophila simulans ]</font>''<font size="2">[http://www.ncbi.nlm.nih.gov/genomes/framik.cgi?db=genome&gi=17601 mitochondria]</font> ''''''<br /> -''<font size="2">[http://www.ncbi.nlm.nih.gov/genomes/framik.cgi?db=genome&gi=116 Arabidopsis thaliana ]</font>''[http://www.ncbi.nlm.nih.gov/genomes/framik.cgi?db=genome&gi=116 <font size="2">mitochondrion</font>]<br />''' | '''NCBI:<br />[http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Info&id=780&lvl=3&lin=f&keep=1&srchmode=1&unlock Taxonomy]<br /> Genome<br /> (examples):<br /> -''<font size="2">[http://www.ncbi.nlm.nih.gov/genomes/framik.cgi?db=genome&gi=17601 Drosophila simulans ]</font>''<font size="2">[http://www.ncbi.nlm.nih.gov/genomes/framik.cgi?db=genome&gi=17601 mitochondria]</font> ''' '''<br /> -''<font size="2">[http://www.ncbi.nlm.nih.gov/genomes/framik.cgi?db=genome&gi=116 Arabidopsis thaliana ]</font>''[http://www.ncbi.nlm.nih.gov/genomes/framik.cgi?db=genome&gi=116 <font size="2">mitochondrion</font>]<br />''' | ||
|} | |} | ||
[[Image:mitochondria.gif|frame|right|Mitochondria. Courtesy of [http://employees.csbsju.edu/hjakubowski/classes/ch331public/ch331.html Dr. Henry Jakubowski.]]] | [[Image:mitochondria.gif|frame|right|Mitochondria. Courtesy of [http://employees.csbsju.edu/hjakubowski/classes/ch331public/ch331.html Dr. Henry Jakubowski.]]] | ||
Revision as of 20:11, 6 June 2006
|
NCBI: |
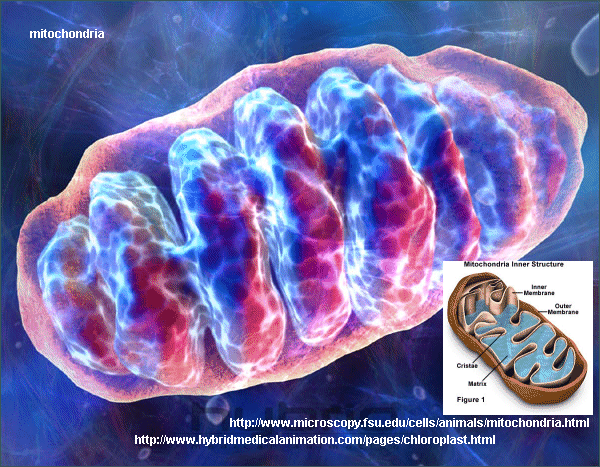
Classification
Higher order taxa:
Bacteria; Proteobacteria; Alphaproteobacteria; Rickettsiales; Rickettsiaceae; Rickettsieae
Species:
Most species of eukaryotes, including plant, fungi, and animal cells contain mitochondria. Mitochondria are phylogenically most closely related to the microbe Rickettsia prowazekii. They are thought to be monophyletic.
Description and Significance
The mitochondrion is an extremely interesting and important organelle in eukaryotic cells. It is the only organelle (other than the nucleus, of course) that has its own DNA independent of the cell's chomosomal DNA; because of this and the fact that the organelle divides independent of the cell, the mitochondrion is thought to have once been a bacterial cell that colonized a eukaryotic cell. Among other things, it performs cellular respiration, has an eletron transport system that occurs across membranes, and produces ATP.
DNA Structure
The mitochondrial DNA is most similar to the genome of Rickettsia prowazekii; both have been described as small, highly derived, and "reduced and tailored to suit its dependent lifestyle " (Gray 1998). The structure of the DNA is double-stranded, circular, and about 16,569 bp in length for human mitochondria. One non coding part of the DNA, called the D-loop, is triple stranded and contains extra 7S DNA. Generally, there are 2 to 10 copies of the DNA in each mitochondrion and many mitochondia in each cell. The mtDNA encodes for 37 genes including 13 mitochondrial peptide subunits, 2 rRNAs, and 22 tRNAs (WUSTL). However, there is a large difference in the limited coding capacity of animal mitochondria and the relatively "large, complicated genomic architectures" of plant mitochondria (Kurland and Andersson 2000).
Structure and Metabolism
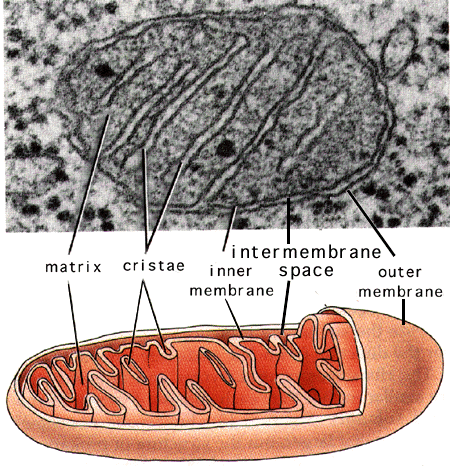
The mitochondrion has four compartments: an outer membrane, an inner membrane (made of cardiolipin), an intermembrane space (between outer and inner membranes), and a matrix (inside inner membrane). The processes that happen in the mitochondron are pyruvate oxidation, the Krebs cycle, the metabolism of amino acids, fatty acids, and steroids, and generation of adenosine triphosphate (ATP). ATP, which is used for energy, is made through the electron-transport chain and the oxidative-phosphoylation system (respiratory chain) in the inner mitochondrial membrane. (WUSTL) View an animated diagram of the proton pump and ATP synthesis from WUSTL. Notice the folds, or cristae, that adds surface area to the mitochondrial inner membrane on the picture and diagram to the right.
Origin
Mitochondria are thought to be aerobic bacterial cells much like Rickettsia bacteria that colonized primordial eukaryotic cells without the ability to use oxygen. Thus, these intracellular aerobic bacteria added oxidative metabolism to the eukaryotic cells and eventually evolved into mitochondria. (WUSTL)
A theory on why a bacterium became an organelle has to do with the increase in ambient oxygen tension in Earth's atmosphere approximately 2 billion years ago. Supposedly, the oxygen tension went from 1% to more than 15% of the current levels within about 200 million years; this "environmental trauma" is thought to have pushed the symbiosis that lead to mitochondria organelles in primitive eukaryotes. (Kurland and Andersson 2000)
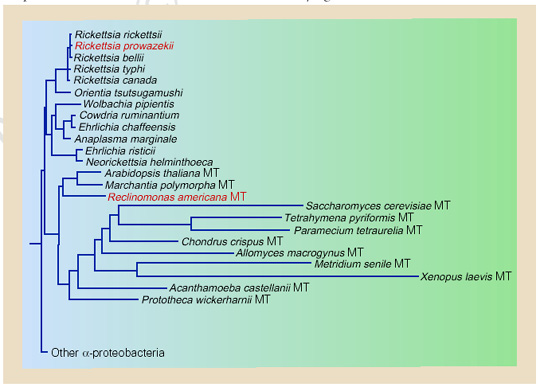
References
General:
- The Mitochondria Research Society
- Dr. Jakubowski: Oxidation/Phosphorylation
- MITOMAP: A Human Mitochondrial Genome Database
- Washington Universtiy, St. Louis: Mitochondrial Disorders
- Andersson, Siv G. E., Alireza Zomorodipour, Jan O. Andersson, Thomas Sicheritz-Ponten, U. Cecilia M. Alsmark, Raf M. Podowski, A. Kristina Naslund, Ann-Sofie Eriksson, Herbert H. Winkler, and Charles G. Kurland. 1998. "The genome sequence of Rickettsia prowazekii and the origin of mitochondria." Nature, vol. 396. Macmillan Publishers Ltd. (133-143)
- Gray, Michael W. 1998. "Rickettsia, typhus and the mitochondrial connection." Nature, vol. 396. Macmillan Publishers Ltd. (109-110)
- Kurland, C. G. and S. G. E. Andersson. 2000. "Origin and evolution of the mitochondrial proteome." Microbiology and Molecular Biology, vol. 64, no. 4. American Society for Microbiology. (786-820)
