Epulopiscium fishelsoni: Difference between revisions
| Line 30: | Line 30: | ||
Include a picture or two (with sources) if you can find them. | Include a picture or two (with sources) if you can find them. | ||
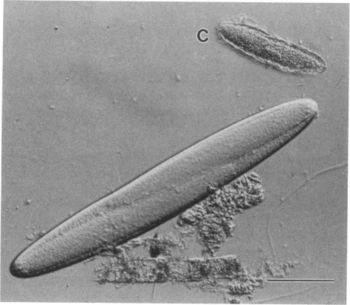
Epulopiscium was first discovered in 1985 by Lev Fishelson. He found them inside the intestines of a brown surgeonfish and was initially classified as a protist because of its large size. ''Epulopiscium fishelsoni'' was later discovered by Linn Montgomery and in 1993 Norman Pace performed an rRNA analysis which confirmed that it was actually a bacteria. At the time ''E. fishelsoni'' was the largest known bacteria, ranging from a length of ~30 to >600μm and a volume of >2,000-fold [2]. ''E. fishelsoni'' is a gram-positive bacterium that has an explicit symbiotic relationship with its surgeonfish hosts. | Epulopiscium was first discovered in 1985 by Lev Fishelson. He found them inside the intestines of a brown surgeonfish and was initially classified as a protist because of its large size. ''Epulopiscium fishelsoni'' was later discovered by Linn Montgomery and in 1993 Norman Pace performed an rRNA analysis which confirmed that it was actually a bacteria. At the time ''E. fishelsoni'' was the largest known bacteria, ranging from a length of ~30 to >600μm and a volume of >2,000-fold. The cell size also varies more than in any other bacteria [2]. ''E. fishelsoni'' is a gram-positive bacterium that has an explicit symbiotic relationship with its surgeonfish hosts. | ||
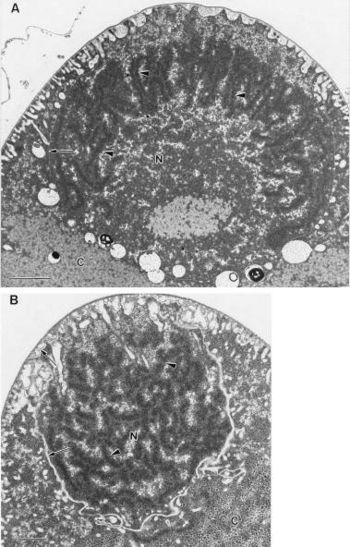
[[Image:Tileshop.fcgi.jpg|thumb|350px|right|Transmission electron micrographs of compact nucleoids of ''E. fishelsoni''. Copyright © 1998, American Society for Microbiology]] | [[Image:Tileshop.fcgi.jpg|thumb|350px|right|Transmission electron micrographs of compact nucleoids of ''E. fishelsoni''. Copyright © 1998, American Society for Microbiology]] | ||
Revision as of 18:46, 29 August 2007
A Microbial Biorealm page on the genus Epulopiscium fishelsoni
Classification
Higher order taxa
Kingdom: Bacteria
Phylum: Firmicutes
Class: Clostridia
Order: Clostridiales
Genus: Epulopiscium
Species
|
NCBI: Taxonomy |
Epulopiscium fishelsoni
Description and significance
Describe the appearance, habitat, etc. of the organism, and why it is important enough to have its genome sequenced. Describe how and where it was isolated. Include a picture or two (with sources) if you can find them.
Epulopiscium was first discovered in 1985 by Lev Fishelson. He found them inside the intestines of a brown surgeonfish and was initially classified as a protist because of its large size. Epulopiscium fishelsoni was later discovered by Linn Montgomery and in 1993 Norman Pace performed an rRNA analysis which confirmed that it was actually a bacteria. At the time E. fishelsoni was the largest known bacteria, ranging from a length of ~30 to >600μm and a volume of >2,000-fold. The cell size also varies more than in any other bacteria [2]. E. fishelsoni is a gram-positive bacterium that has an explicit symbiotic relationship with its surgeonfish hosts.
Genome structure
Describe the size and content of the genome. How many chromosomes? Circular or linear? Other interesting features? What is known about its sequence? Does it have any plasmids? Are they important to the organism's lifestyle?
Cell structure and metabolism
Describe any interesting features and/or cell structures; how it gains energy; what important molecules it produces.
E. fishelsoni has a layer of flagella around its surface. When viewed in a light microscope video the flagella layer is vibrating in a wave-like motion which is what makes the cell swim. Beneath the outer surface is a peripheral layer of rolled up membranes, followed by a dense region with large, lightly stained inclusions. In the central region of the cell there are often membrane-bound daughter cells that is involved in the reproductive process. Upon completion of development, the daughter cells emerge through a centrally located, oblong split in the envelope of the mother cell. Epulos possess bacterial-type flagella rather than eukaryote-type cilia. The epulo flagella have a diameter of 14 to 18 nm in thin section and in freeze fracture profile, compared with 200 to 250 nm for eukaryote cilia. It has not been possible to negatively stain the flagella of very large epulo specimens.
Epulos possess bacterial-type nucleoids rather than eukaryote-type nuclei. The nucleoid DNA is found in circumscribed regions scattered throughout the mother cell cytoplasm, while in the daughter cells it is often found as a concentric peripheral region. Its peripheral distribution is atypical for bacteria (11, 20) but may be related to the maturation of the daughter cell. It has a coagulated appearance (Fig. 6) typical of that seen in the bacterial nucleoid after standard fixation for electron microscopy (11, 20). The nucleoids do not have a surrounding membrane. The central region of the daughter cell contains many ribosomes, typical of bacterial cytoplasm in this regard, although its appearance is more heterogeneous than is usually seen. The limiting membranes of the daughter-cell do not show any structures resembling nuclear pores
Ecology
Describe any interactions with other organisms (included eukaryotes), contributions to the environment, effect on environment, etc.
Pathology
E. fishelsoni is not known to cause any disease or be harmful to anything. It has a symbiotic relationship with surgeonfish which seems to be mutually beneficial. In fact, E. fishelsoni can not be cultured outside of its host and can only be found in the gut of that family of fish.
Application to Biotechnology
E. fishelsoni does not produce any useful compound or enzymes that can be used for biotechnology. All that is known is that this bacteria helps regulate the pH in the gut of its host. This regulation is cycled throughout the day and is controlled based on the surgeonfish's activity.
Current Research
There are currently no ongoing nor recent research being done about E. fishelsoni. After its discovery in 1985, thorough examinations of its structure and research about its reproduction process in the 90's, there is not much left that we don't know about this bacteria. There aren't any signs nor indications of future research projects for E. fishelsoni.
References
Edited by student of Rachel Larsen