Desulfobacter: Difference between revisions
| Line 17: | Line 17: | ||
'''NCBI: [http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Info&id=2289&lvl=3&lin=f&keep=1&srchmode=1&unlock Taxonomy] Genome''' | '''NCBI: [http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Info&id=2289&lvl=3&lin=f&keep=1&srchmode=1&unlock Taxonomy] Genome''' | ||
|} | |} | ||
''D. postgatei'', ''D. hydrogenophilus'', ''D. | |||
latus'', and ''D. curvatus'' | |||
==Description and Significance== | ==Description and Significance== | ||
Revision as of 07:34, 17 March 2008
A Microbial Biorealm page on the genus Desulfobacter
[[Image:.jpg|thumb|350px|right|
Classification
Higher order taxa:
Bacteria; Proteobacteria; delta/epsilon subdivisions; Deltaproteobacteria; Desulfobacterales; Desulfobacteraceae
Species:
|
NCBI: Taxonomy Genome |
D. postgatei, D. hydrogenophilus, D. latus, and D. curvatus
Description and Significance
Desulfobacter is a mesophilic gram-negative genera that is capable of oxidizing organic substrate completely to CO2 in anoxic codition. Desulfobacter is mainly found in brackish and marine environemnt and has a oval or vibrio morphology [1]. The genus desulfobacter includes acetateutelizing sulphate-reducing bacteria. 10 Methyl 16:0 is an unusual bacterial phospholipid fatty acids (PLFA) characteristic of Desulfobacter-type sulphate-reducing bacteria and could be used as a biomarker for these bacteria [2,3].
Genome Structure
Cell Structure and Metabolism
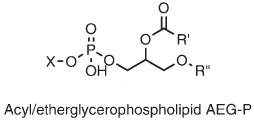
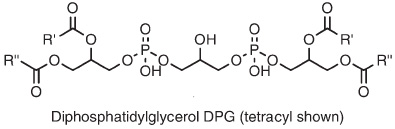
D. variabilis contains significant quantities of AEG-P (top), and almost 20% of the alkyl-glycerol bond is present as DPG lipids, also known as "cardiolipin" (bottom). Note that the DPGs contain either ether or ester linkages in the core lipids.Sturt et al |
| Sulfate reducers have a wide range of cellular morphologies, including rods, vibrios, ovals, spheres and even tear-dropped or onion shaped cells. Some are motile, others are not. Most sulfate-reducing bacteria are mesophilic, but a few thermophiles are known. Desulfosarcina variabilis is mesophilic, and contains bacterial core lipids (see images on left). The dominant phospholipid headgroups in D. variabilis are Phosphoethanolamine PE (48%) and Phosphoglycerol PG (33%). One study has found that Desulfosarcina variabilis solely contained n-hexadecyl ether side chains. For more information on tetraether lipids found in Archaea, click here. |
Ecology
Micobiological and molecular studies have suggested the presence of different sulfate reducing bacteria (SBR)in oxic and anoxic layers of soil profile. Dsulfobater, Desulfobulbus and desulfotomoculum species were detected in restricly anoxic conition while Desulfococcus, Desolfonema, and Desulfovibrio species appear to be perdominantly in oxic layers [4]. The main habitat of Desulfobacter is marine sendiment and brackish water.

References
Arches, specifically p.10 populations and methods.PDF here.
[http://www.bact.wisc.edu/Microtextbook/modules.php?op=modload&name=Sections&file=index&req=viewarticle&artid=104&page=1 Microbiology Textbook. Copyright, Timothy Paustian© 1999-2004.]
[1] Lary L. Barton Biotechnology handbooks. Vol. 8-Sulfate reducing bacteria. 1995 Plenum Press, New York.
[2] Dowling, N.J., Widdel, F. and White, D.C. (1986) Comparison of the phospholipid ester-linked fatty acid biomarkers of acetate-oxidising sulphate-reducers and other sulphide-forming bacteria. J. Gen. Microbiol. 132,1815-1825.
[3] Dowling, N.J., Nichols, P.D. and White, D.C. (1988)Phospholipid fatty acid and infra-red spectroscopic analysis of a sulfate-reducing consortium. FEMS Microbiol.Ecol. 53, 325-334.
[4] Sulphate reducing bacteria-book)