Papillomaviridae: Difference between revisions
No edit summary |
|||
| Line 10: | Line 10: | ||
==Description and Significance:== | ==Description and Significance:== | ||
Papillomaviruses are a class of viruses that infect many vertebrates and can cause benign epithelial growths, known as papillomas, such as skin and genital warts. | |||
Human Papilloma Virus (HPV) is a very common virus that causes the growth of abnormal tissue or cells on body skin. HPV can cause abnormal tissue changes on the vocal cords, mouth, hands, feet and genital organs. Each type of HPV infects certain parts of the body and over 60 such types have already been identified. | Human Papilloma Virus (HPV) is a very common virus that causes the growth of abnormal tissue or cells on body skin. HPV can cause abnormal tissue changes on the vocal cords, mouth, hands, feet and genital organs. Each type of HPV infects certain parts of the body and over 60 such types have already been identified. | ||
Revision as of 20:30, 21 June 2006
Baltimore Classification
Higher order taxa
root; Viruses; dsDNA viruses, no RNA stage; Papillomaviridae; Papillomavirus
Species
Human papillomavirus, Bovin papillomavirus, Fringilla coelebs papillomavirus (examples)
Description and Significance:
Papillomaviruses are a class of viruses that infect many vertebrates and can cause benign epithelial growths, known as papillomas, such as skin and genital warts.
Human Papilloma Virus (HPV) is a very common virus that causes the growth of abnormal tissue or cells on body skin. HPV can cause abnormal tissue changes on the vocal cords, mouth, hands, feet and genital organs. Each type of HPV infects certain parts of the body and over 60 such types have already been identified.
HPV is of extreme significance because some types of this virus lead to the growth of abnormal tissues that can lead to cancer of the female organs. Cancer can be prevented by finding and treating HPV-related tissue changes.
Genome Structure
Papillomaviruses are a group of small non-enveloped DNA tumor viruses whose infection usually causes benign epithelial lesions (warts). Certain types of HPVs, such as HPV-16, HPV-18, and HPV-31 have been recognized as causative agents of cervical cancer and anal cancer. Sexual transmission cause their transmission and are associated with more than 95% of cervical cancer. Papillomaviruses infect keratinocytes in the basal layer of stratified squamous epithelia and replicate in the nucleus of infected keratinocytes in a differentiation-dependent manner.
The papillomavirus genome consists of circular, d/s DNA ~8kbp in size, associated with cellular histones to form a chromatin-like substance. At least 12 different HPV genomes have been sequenced, and the genetic organization of all is similar. (source: Microbiology@Leicester: Papillomaviruses)
Virion Structure of a Papillomavirus
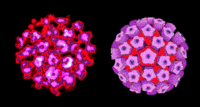
Papillomaviruses are small, non-enveloped icosahedral particles, approximately 51-55nm in diameter. 72 morphological units, or capsomers, are arranged on the surface of a T=7 icosahedran to form a capsid. The capsomers located at each of the 12 vertices are pentavalent--each capsomer is surrounded by five adjecent capsomers-- and the other 60 capsomers are hexavalent-- each capsomer is adjacent to six capsomers. There is one major capsid protein encoded by the L1 gene and one minor capsid protein encoded by the gne L2. (source: Microbiology@Leicester: Papillomaviruses)
Reproductive Cycle of a Papillomavirus in a Host Cell
The individual isolates for Papillomavirus are very species-specific. All of these isolates are tropic for squamous epithelial cells but the receptors are unknown. The virus infects the basal cells of the dermal layer, and early gene expression can be detected in these cells (in situ hybridization). However, late gene expression, expression of structural proteins and vegetative DNA synthesis is restricted to terminally differentiated cells of the epidermis which implies a link between cellular differentiation and viral gene expression.
The functions of each Gene are given below:
E1-- DNA-dependent ATPase, ATP dependent helicase: allow unwinding of the viral genome and act as an elongation factor for DNA replication.
E2-- Responsible for recognition and binding of origin of replication. Exists in two forms: full length (transcriptional transactivator) and truncated (transcriptional repressor). The ratio of these found in the heterotrimeric complex formed before complexing with E1 regulates transcription of viral genome.
E3-- Not known
E4-- Late Expression: C terminal binds intermediate filament, allowing release of virus-like particles. Also involved in transformation of host cell by deregulation of host cell mitogenic signalling pathway.
E5-- Obstruction of growth suppression mechanisms: e.g EGF receptor; activation of mitogenic signalling pathways via transcription factors: c-Jun and c-Fos (important in ubiquitin pathway degradation of p53 complex by E6). Inactivation of p21 (p53 induced expression halts cell cycle until DNA is proof-read for mutations).
E6-- E6 Transformation of host cell by binding p53 tumour suppressor protein.
E7-- Transforming protein, binds to pRB/p107.
E8-- Not known
L1-- Major capsid protein: can form virus-like particles.
L2-- Minor capsid protein: possible DNA packaging protein.
The expression of the Papillomavirus genome is complex because there are multiple promoters, alternative splicing patterns, and a link between differentiation and gene expression. Only one strand of the genome is transcribed, producing two classes of proteins: the non-structural regulatory proteins, known as early proteins, and the structural proteins L1 and L2, known as late proteins.
The transformation process is very complex and depends largely on the early gene products. The transferring proteins appear to vary from one virus type to another and the function and mechanism of these transforming proteins are still not clear. The general principle appears to be that two or more early proteins co-operate to give a transforming phenotype.The most confusing thing is that in most cases, all or part of the Papilloma genome including the putative "transforming genes" is maintained in the tumour cells, whereas in other cases, the virus DNA may be lost after transformation. BPV-4 is an example of one such "hit-and-run" mechanism.
In case of Human Papillomaviruses (HPVs), E2 binds to the early promoter and decreases expression of E6/E7. The loss of E2 is thus the first stage of transformation. E6 then binds to p53 via a cellular protein, p100, and targets it for degradation via the ubiquitin pathway. E7 binds pRB and prevents phosphorylation. This would normally result in apoptosis but both E6 ad E7 interact with a number of cellular proteins which influence the outcome of infection.
The HPV E7 proteins are small (HPV16 E7 comprising 98 amino acids), zinc binding phosphoproteins which are localised in the nucleus. They are structurally and functionally similar to the E1A protein of subgenus C adenoviruses. The first 16 amino-terminal amino acids of HPV16 E7 contain a region homologous to a segment of the conserved region 1 (CR1) of the E1A protein of subgenus C adenoviruses. The next domain, up to amino acid 37, is homologous to the entire region 2 (CR2) of E1A. Genetic studies have established that these domains are required for cell transformation in vitro, suggesting similarities in the mechanism of transformation by these viruses. The CR2 homology region contains the LXCXE motif (residues 22-26) involved in binding to the tumour suppressor protein pRb. This sequence is also present in SV40 and polyoma large T antigens. The high risk HPV E7 proteins (of, for example, types 16 and 18) have an approximately ten-fold higher affinity for pRb protein than the low risk HPV E7 proteins (of, for example, type 6). Association of the E7 protein with pRb promotes cell proliferation by the same mechanism as the E1A proteins of adenoviruses and SV40 large T antigen. Recent studies have shown that E7 promotes degradation of Rb family proteins rather than simply inhibiting their function by complex formation. The CR2 region also contains the casein kinase II phosphorylation site (residues 31 and 32). The remaining 61 amino acids of E7 protein have very little similarity to E1A, however a sequence CXXC involved in zinc binding is present in both proteins. The E7 protein contains two of these motifs which mediate dimerisation of the protein. Mutation in one of the two Zn binding motifs destroys transforming activity, although this mutant is able to associate with Rb protein. Therefore dimerisation may be important for the transforming activity of E7.
The HPV E6 are small basic proteins (HPV16 E6 comprising 151 amino acids) which are localised to the nuclear matrix and non-nuclear membrane fraction. They contain four cysteine motifs which are thought to be involved in zinc binding. E6 encoded by high risk HPVs associates with the wild type p53 tumour suppressor protein. For association with p53, the E6 protein requires a cellular protein of 100 kDa, termed E6-associated protein (E6-AP). Like SV40 large T antigen and Ad5 E1B 58 kDa, E6 proteins of high risk HPVs abrogate the ability of wild type p53 to activate transcription. However, the mechanism of E6 action is different than that of SV40 large T and the E1B protein since it involves degradation of p53. It has been shown that E6-dependent degradation of p53 occurs through the cellular ubiquitin proteolysis pathway.
Genome Replication: The genome is replicated as a multicopy nuclear plasmid (episome). There are two mechanisms involved in genome replication:
Plasmid Replication involves the E1 protein and occurs in cells in the lower levels of the dermis. The virus DNA is amplified to 50-400 diploid genomes and then it replicates once per cell division, with the copy number/cell remaining constant.
Vegetative Replication occurs in terminally differentiated cells in the epidermis. Here, the control over the number of copies seems to be lost and the DNA is amplified to hundreds of copies/cell. The virus is shed from epidermal cells when these are sloughed off and is transmitted by both direct and indirect contact.
The differentiation of the host cell determines the productive infection by HPV. A low level of gene expression occurs when HPV genomes are established as autonomous replicating extrachromosomal elements following entry into basal epithelial cells. After the differentiation of infected cells, the synthesis of progeny virions is made possible because of the induction of productive replication and expression of caspid genes. Certain HPV types can also exist in a latent state, as exemplified by evidences from immunosupressed patients as well as individuals with recurring laryngeal papillomatis.While HPV DNA may be present, differentiation-dependent synthesis of virions is not possible in latently infected cells. The effectiveness of therapeutic methods for treatment of infections could be determined by the presence of a latent state for HPVs. (source: Microbiology@Leicester: Papillomaviruses)
Viral Ecology & Pathology
HPV can grow on the cervix, vagina, vulva, urethra and the anus. HPV causes condyloma (warts) and dysplasia (pre-cancer), two kinds of abnormal tissues. There is very little information available as to how or when people become infected with HPV. A lot of medical research is in progress to answer these questions and understand the exact nature of HPVs. It is only known now that HPV is mainly spread through sexual contact. The symtoms for HPV are not very explicit and both men and women infected with the virus may not know about it, nor develop dysplasia or genital warts for many years. Interestingly, smoking ciggarettes is also closely associated to HPV with smokers having a much higher chance of developing dysplasia than non-smokers.
References
Virus Alpha Structure; Electron Micrograph Images: Papillomavirus
The Big Picture Book of Viruses: Papovaviridae
Department of Obstetrics and Gynecology: Human Papillomavirus
Papillomavirus genome structure, expression, and post-transcriptional regulation
