Dictiostelida: Difference between revisions
No edit summary |
No edit summary |
||
| Line 1: | Line 1: | ||
{{Biorealm}} | |||
{| | {| | ||
| height="69" bgcolor="#FFDF95" | | | height="69" bgcolor="#FFDF95" | | ||
Revision as of 14:56, 14 August 2006
A Microbial Biorealm page on the Dictiostelida
|
NCBI: |
Classification
Higher order taxa:
Eukaryota; Mycetozoa
Species:
Dictyostelium discoideum, D. firmibasis, D. giganteus
Description and Significance
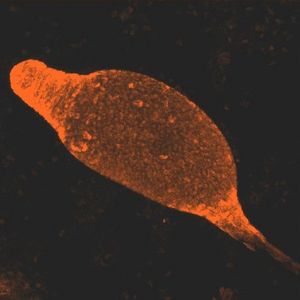
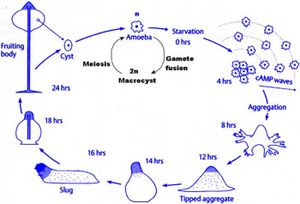
Dictyostelium are known as cellular slime molds. They differ from plasmodial slime molds because plasmodial slime molds are a multinucleated amoeboid mass, not an aggregation of individual cells. Dictyostelium resemble fungi in many respects, but are classified as protists. Dictyostelium were first discovered in North Carolina in 1935. Dictyostelium are social amebas, meaning that they live as both unicellular and multicellular organisms. Dictyostelium start out as separate, individual cells. In the event of adverse conditions, such as starvation, up to 100,000 cells will signal each other with cAMP and converge via chemotaxis. This process takes about 12 hours. This mass of cells is surrounded by an extracelluar matrix. This multicellular structure resembles a slug. This slug leave a trail of slime as it migrates. Some claim that the slug stage is a single organism, while others claim that it is a colony.
Genome Structure
The genome of the species Dictyostelium discoideum was mapped by Eichinger et. al. (2005). The six chromosomes of this species are gene-dense. It is estimated that this species contains 8,000 to 10,000 genes. Studies of known genes indicate a high degree of sequence similarity to genes in vertebrate species. This suggests that D. discoideum is close to the base of metazoan evolution. They found approximately 12,500 predicted protiens. There are many complex repeats in this genome. Eichinger et. al. found fragments of extrachromosomal ribosomal DNA (rDNA) at the ends of each chromosome. This suggests two things: a novel telomere structure, and the use of common mechanisms to maintain rDNA and chromosomal termini.
Cell Structure and Metabolism
As social amebas, Dictyostelium are able to alternate between unicellular and multicelluar forms. There are no cell walls on these organisms. Individual cells are not flagellated, and thus are largely non-motile. These individual cells eventually aggregate by sliding. The cytoplasm projects to form a pseudopod, allowing the organism to travel. The work of Saito et. al. (2005) illustrates one of the ways in which Dictyostelium adapts to changes in environment. In response to elevated temperatures, Dictyostelium change their fatty acid composition to maintain membrane fluidity.
Dictyosteilum feed on bacteria; they are heterotrophic. They are considered engulfers, meaning they engulf their prey.
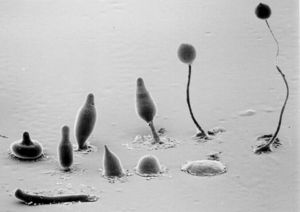
Dictyostelium are both haploid and diploid organisms. Sexual and asexual reproduction are both options. The transition from unicellular organism to multicellular fruiting body makes up an important part of Dictyostelium life cycles. After aggregation occurs, the amoebae differentiate between prestalk and prespore cells. These eventually form the stalk and the spore head. Prestalk cells form at the top of the organism and form the tip. This tip controls further development and cell movement. Finally, a fruiting body forms, with spores at the top of the stalk. These spores are dispersed by the wind, and release amoebae. The work of Gundersen et. al. (2005) shows the importance of the G alpha2 subunit on Dictyostelium life cycles. G alpha2 is the alpha subunit for guanine nucleotide binding regulatory proteins. If G alpha2 does not function, cells do not enter the developmental phase.
Ecology
Dictyostelium often form on fruiting bodies with spores, but are usually found inhabiting soil, animal dung, humus, and other decaying matter. They are usually located in forest habitats, and can be found around the world.
Dictyostelium are useful organisms for research. They are ideal for studying cell motility and transduction, cell-type differentiation, and developlemental processes. Dictyostelium discoideum was used by Graf et. al. (2004) in their study of centrosomes.
References
Abedon, Stephen T. "Kingdom" Mycetozoa.
Caprette, David R. Cellular "Slime Molds": Dictyostelium and relatives.
The Dictyostelium Genome Sequencing Project.
Siegert, Florian. Pattern Formation in Dictyostelium discoideum.
