Nitrate/nitrite methane oxidation: Difference between revisions
No edit summary |
No edit summary |
||
| Line 4: | Line 4: | ||
Methane oxidation can occur in oxic and anoxic sediments. Aerobic metabolism of methane requires oxygen so that the methane molecule can be activated. The reaction with oxygen forms methanol, which is a better electron donor than methane. A series of oxidation reactions, where each reaction utilizes a different enzyme, results in the production of CO<sub>2</sub>. <i>Methylococcus capsulatus</i> is the most well-studied aerobic methanotroph (Oremland, 2010). | Methane oxidation can occur in oxic and anoxic sediments. Aerobic metabolism of methane requires oxygen so that the methane molecule can be activated. The reaction with oxygen forms methanol, which is a better electron donor than methane. A series of oxidation reactions, where each reaction utilizes a different enzyme, results in the production of CO<sub>2</sub>. <i>Methylococcus capsulatus</i> is the most well-studied aerobic methanotroph (Oremland, 2010). | ||
There are two known types of anaerobic oxidation of methane. One of the types of anaerobic oxidation entails the interaction of sulfate-reducing bacteria with reverse methanogens in marine sediments. It is thought that methane oxidation fuels sulfate reduction by providing H<sub>2</sub> that sulfate-reducing bacteria can use as an electron donor. Sulfate reduction, in turn, creates thermodynamic conditions that allow for methane oxidation to occur (Valentine, 2000); methane cannot be oxidized in unfavorable thermodynamic conditions because it is a poor electron donor. This symbiotic relationship is summarized as: | There are two known types of anaerobic oxidation of methane. One of the types of anaerobic oxidation entails the interaction of sulfate-reducing bacteria with reverse methanogens in marine sediments. It is thought that methane oxidation fuels sulfate reduction by providing H<sub>2</sub> that sulfate-reducing bacteria can use as an electron donor. Sulfate reduction, in turn, creates thermodynamic conditions that allow for methane oxidation to occur (Valentine and Reeburgh, 2000); methane cannot be oxidized in unfavorable thermodynamic conditions because it is a poor electron donor. This symbiotic relationship is summarized as: | ||
CH<sub>4</sub> + SO<sub>4</sub><sup>2-</sup> --> HCO<sup>3-</sup> + HS<sup>-</sup> + H<sub>2</sub>O | CH<sub>4</sub> + SO<sub>4</sub><sup>2-</sup> --> HCO<sup>3-</sup> + HS<sup>-</sup> + H<sub>2</sub>O | ||
| Line 42: | Line 42: | ||
The metagenomes of two cultures of <i>M. oxyfera</i> from enriched freshwater sediments (collected from Australia and the Netherlands) were sequenced to piece together the mechanism of nitrite-dependent anaerobic oxidation of methane (Ettwig et al, 2010). The researchers found that <i>M. oxyfera</i> expressed genes for the conversion of NO<sup>3-</sup> to NO<sup>2-</sup>, NO<sup>2-</sup> to NO (nitric oxide) and NO to N<sub>2</sub>O (nitrous oxide), but no genes for the conversion of N<sub>2</sub>O to N<sub>2</sub> (Figure 3a). Despite this, N<sub>2</sub>O could only be found in trace amounts. This suggests that the NO is being converted to N<sub>2</sub> using an alternative mechanism where N<sub>2</sub>O is not an intermediate. On the other hand, it may be that N<sub>2</sub>O is being reduced to N<sub>2</sub> using, not N<sub>2</sub>O reductase, but a different enzyme. | The metagenomes of two cultures of <i>M. oxyfera</i> from enriched freshwater sediments (collected from Australia and the Netherlands) were sequenced to piece together the mechanism of nitrite-dependent anaerobic oxidation of methane (Ettwig et al, 2010). The researchers found that <i>M. oxyfera</i> expressed genes for the conversion of NO<sup>3-</sup> to NO<sup>2-</sup>, NO<sup>2-</sup> to NO (nitric oxide) and NO to N<sub>2</sub>O (nitrous oxide), but no genes for the conversion of N<sub>2</sub>O to N<sub>2</sub> (Figure 3a). Despite this, N<sub>2</sub>O could only be found in trace amounts. This suggests that the NO is being converted to N<sub>2</sub> using an alternative mechanism where N<sub>2</sub>O is not an intermediate. On the other hand, it may be that N<sub>2</sub>O is being reduced to N<sub>2</sub> using, not N<sub>2</sub>O reductase, but a different enzyme. | ||
Ettwig et al (2010) also found that the metagenome did not contain genes coding fumarate-adding glycl-radical enzymes or methyl-coenzyme M reductase, either of which can be used for anaerobic oxidation of methane. Rather, they found that the metagenome coded for the aerobic oxidation of methane (Figure 3b). First, methane reacts with oxygen, using the membrane-bound form of the enzyme methane mono-oxygenase (pMMO), to form methanol (CH<sub>3</sub>OH) and water. The pMMO complex is thought to function either by aiding oxygen production and then consuming the oxygen produced or by using NO directly as an oxygen source. Further research needs to be conducted to establish the mechanism by which pMMO works. | Ettwig et al (2010) also found that the metagenome did not contain genes coding fumarate-adding glycl-radical enzymes or methyl-coenzyme M reductase, either of which can be used for anaerobic oxidation of methane. Rather, they found that the metagenome coded for the aerobic oxidation of methane (Figure 3b). First, methane reacts with oxygen, using the membrane-bound form of the enzyme methane mono-oxygenase (pMMO), to form methanol (CH<sub>3</sub>OH) and water. The pMMO complex is thought to function either by aiding oxygen production and then consuming the oxygen produced or by using NO directly as an oxygen source. Further research needs to be conducted to establish the mechanism by which pMMO works in <i>M. oxyfera</i>. | ||
A separate enzyme, methanol dehydrogenase, is used to form formaldehyde (CH<sub>2</sub>O) from CH<sub>3</sub>OH. CH<sub>2</sub>O, in turn, is oxidized to form formate (HCOO<sup>-</sup>). The conversion of CH<sub>2</sub>O to HCOO<sup>-</sup> requires a tetrahydromethanopterin-dependent C1 transfer module which is thought to be a primordial metabolic molecule; this indicates that <i>M. oxyfera</i> is from a particularly deep-branching lineage. Finally, HCOO<sup>-</sup> is converted to CO<sub>2</sub> using the enzyme formate dehydrogenase. The electron flow resulting from the oxidation of methane generates a proton motive force and therefore allows for ATP synthesis. | A separate enzyme, methanol dehydrogenase, is used to form formaldehyde (CH<sub>2</sub>O) from CH<sub>3</sub>OH. CH<sub>2</sub>O, in turn, is oxidized to form formate (HCOO<sup>-</sup>). The conversion of CH<sub>2</sub>O to HCOO<sup>-</sup> requires a tetrahydromethanopterin-dependent C1 transfer module which is thought to be a primordial metabolic molecule; this indicates that <i>M. oxyfera</i> is from a particularly deep-branching lineage. Finally, HCOO<sup>-</sup> is converted to CO<sub>2</sub> using the enzyme formate dehydrogenase. The electron flow resulting from the oxidation of methane generates a proton motive force and therefore allows for ATP synthesis (Ettwig et al, 2010). | ||
[[Image:Kanmani_figure_4.jpg|thumb|700px|right|Figure 4. Nitrite-dependent methane oxidation in cultures of <i>M. oxyfera</i> overtime. Nitrite was labeled with <sup>15</sup>N. Arrows indicate addition of labeled nitrite. Red circles, CH<sub>4</sub>; dark blue triangles, <sup>15,15</sup>N<sub>2</sub>; light blue triangles, <sup>15,14</sup>N<sub>2</sub>; green squares, total N<sub>2</sub>; dark green squares, <sup>14,15</sup>N<sub>2</sub>O and <sup>15,15</sup>N<sub>2</sub>O. (a) Addition of labeled nitrite is associated with a decrease in CH<sub>4</sub> and increase in labeled and partly labeled N<sub>2</sub>. (b) Addition of labeled nitrite does not consume or produce N<sub>2</sub>O.]] | [[Image:Kanmani_figure_4.jpg|thumb|700px|right|Figure 4. Nitrite-dependent methane oxidation in cultures of <i>M. oxyfera</i> overtime. Nitrite was labeled with <sup>15</sup>N. Arrows indicate addition of labeled nitrite. Red circles, CH<sub>4</sub>; dark blue triangles, <sup>15,15</sup>N<sub>2</sub>; light blue triangles, <sup>15,14</sup>N<sub>2</sub>; green squares, total N<sub>2</sub>; dark green squares, <sup>14,15</sup>N<sub>2</sub>O and <sup>15,15</sup>N<sub>2</sub>O. (a) Addition of labeled nitrite is associated with a decrease in CH<sub>4</sub> and increase in labeled and partly labeled N<sub>2</sub>. (b) Addition of labeled nitrite does not consume or produce N<sub>2</sub>O (Ettwig et al, 2010).]] | ||
A peculiar aspect of the pathway is that nitrite is required for the mechanism to proceed even though <i>M. oxyfera</i> contains genes that code nitrate reductase (Figure 3a). Ettwig et al (2010) conducted an experiment where they provided an enrichment culture, under anoxic conditions, with nitrate and carbon-13 labeled CH<sub>4</sub>. Methane oxidation did not occur and N<sub>2</sub> gas was not produced when the culture was incubated. However, as soon as nitrogen-15 labeled nitrite was added, methane oxidation commenced and labeled N<sub>2</sub> gas was formed in stoichiometric quantities, indicating that N<sub>2</sub> was produced solely from the added nitrite (Figure 4a). Is nitrate reductase not produced even though the metagenome codes for it? | A peculiar aspect of the pathway is that nitrite is required for the mechanism to proceed even though <i>M. oxyfera</i> contains genes that code nitrate reductase (Figure 3a). Ettwig et al (2010) conducted an experiment where they provided an enrichment culture, under anoxic conditions, with nitrate and carbon-13 labeled CH<sub>4</sub>. Methane oxidation did not occur and N<sub>2</sub> gas was not produced when the culture was incubated. However, as soon as nitrogen-15 labeled nitrite was added, methane oxidation commenced and labeled N<sub>2</sub> gas was formed in stoichiometric quantities, indicating that N<sub>2</sub> was produced solely from the added nitrite (Figure 4a). Is nitrate reductase not produced even though the metagenome codes for it? | ||
| Line 68: | Line 68: | ||
<br>The global budget of methane in the atmosphere is 500-600 Tg CH<sub>4</sub> per year. Its levels have increased between pre-industrial times and now from 715 ppbv to 1770 ppbv. Much of this increase can be attributed to fossil fuel burning and the intensification of agriculture – ruminants and large-scale rice plantation produce substantial amounts of methane (Conrad, 2009). The increase of methane in the atmosphere has significantly contributed to climate change, given its status as a greenhouse gas with a much higher global warming potential that carbon dioxide. | <br>The global budget of methane in the atmosphere is 500-600 Tg CH<sub>4</sub> per year. Its levels have increased between pre-industrial times and now from 715 ppbv to 1770 ppbv. Much of this increase can be attributed to fossil fuel burning and the intensification of agriculture – ruminants and large-scale rice plantation produce substantial amounts of methane (Conrad, 2009). The increase of methane in the atmosphere has significantly contributed to climate change, given its status as a greenhouse gas with a much higher global warming potential that carbon dioxide. | ||
Climate warming has a positive feedback loop. In the case of wetlands, for example, warming has accelerated the actions of soil methanogens, thereby increasing the release of methane from the soil. Since methane is a potent greenhouse gas, the increase in methane levels will only increase warming. This, in turn will further increase methane release from the soil provided that soil moisture levels remain high (Cao et | Climate warming has a positive feedback loop. In the case of wetlands, for example, warming has accelerated the actions of soil methanogens, thereby increasing the release of methane from the soil. Since methane is a potent greenhouse gas, the increase in methane levels will only increase warming. This, in turn will further increase methane release from the soil provided that soil moisture levels remain high (Cao et al, 1998). | ||
Methane-oxidizing bacteria, as mentioned earlier, can mediate methane fluxes and nitrate levels. It may, therefore, be possible to use them for bioremediation. Nitrate/nitrite-dependent methane-oxidizing microbes can be used to offset increases in methane production (Raghoebarsing et al, 2006). In the case of agricultural run-off, the same microbes can be used to consume the nitrate/nitrite in soils and aquatic and marine sediments. These microbes can also be used to restore eutrophic marine and freshwater ecosystems as they thrive in anoxic conditions where there is a lot of nitrite and methane available. The use of the intra-aerobic pathway may help re-oxygenate the ecosystem, allowing the ecosystem to sustain life once again. | Methane-oxidizing bacteria, as mentioned earlier, can mediate methane fluxes and nitrate levels. It may, therefore, be possible to use them for bioremediation. Nitrate/nitrite-dependent methane-oxidizing microbes can be used to offset increases in methane production (Raghoebarsing et al, 2006). In the case of agricultural run-off, the same microbes can be used to consume the nitrate/nitrite in soils and aquatic and marine sediments. These microbes can also be used to restore eutrophic marine and freshwater ecosystems as they thrive in anoxic conditions where there is a lot of nitrite and methane available. The use of the intra-aerobic pathway may help re-oxygenate the ecosystem, allowing the ecosystem to sustain life once again. | ||
Revision as of 02:29, 26 April 2011
Introduction
Methane (CH4) can be oxidized to carbon dioxide (CO2) by microbes in both marine and freshwater sediments. In oceans, methane seeps out of fissures along the ocean floor and from methane hydrates. The methane eventually bubbles out of the ocean and enters the atmosphere. It is estimated approximately 33 million tons of methane enters the atmosphere in this manner per year (Biello, 2006). Methane is also released from freshwater sediments (i.e. from lakes and wetlands) as a result of methanogenic activity. The ability of microbes to degrade methane means that there is a methane sink which can mediate fluxes of methane into the atmosphere. Methane oxidation is an important topic of study given that methane is a potent greenhouse gas. Although methane degradation results in greater CO2 flux into the atmosphere, CO2 has less impact than methane as a greenhouse gas. Bacteria and archaea that can metabolize methane are called methanotrophs and reverse methanogens, respectively.
Methane oxidation can occur in oxic and anoxic sediments. Aerobic metabolism of methane requires oxygen so that the methane molecule can be activated. The reaction with oxygen forms methanol, which is a better electron donor than methane. A series of oxidation reactions, where each reaction utilizes a different enzyme, results in the production of CO2. Methylococcus capsulatus is the most well-studied aerobic methanotroph (Oremland, 2010).
There are two known types of anaerobic oxidation of methane. One of the types of anaerobic oxidation entails the interaction of sulfate-reducing bacteria with reverse methanogens in marine sediments. It is thought that methane oxidation fuels sulfate reduction by providing H2 that sulfate-reducing bacteria can use as an electron donor. Sulfate reduction, in turn, creates thermodynamic conditions that allow for methane oxidation to occur (Valentine and Reeburgh, 2000); methane cannot be oxidized in unfavorable thermodynamic conditions because it is a poor electron donor. This symbiotic relationship is summarized as:
CH4 + SO42- --> HCO3- + HS- + H2O
The other type of anaerobic oxidation is microbial nitrate/nitrite oxidation of methane. This entails the coupling of denitrification to methane oxidation to form CO2. This has become an important topic of research given the intensification of agriculture and the resulting increase in agricultural runoff (Raghoebarsing et al, 2006). In addition, Nauhaus et al (2005) and Raghoebarsing et al (2006) found that nitrate and nitrite were better substrates for anaerobic oxidation of methane than sulfate. This suggests that nitrate/nitrite-dependent methane oxidation may be more prevalent in freshwater and marine ecosystems than sulfate-dependent methane oxidation.
There are two main theories that have been proposed for microbial nitrate/nitrite oxidation of methane. The first theory proposes that a microbial consortium, including bacteria and archaea, is required for the coupling of both processes to occur (Raghoebarsing et al, 2006). The second theory proposes that methane oxidation can occur in the absence of archaea. The bacterium responsible for this has been named Candidatus Methylomirabolis oxyfera, or M. oxyfera. The overall pathway utilized by M. oxyfera has been termed as ‘intra-aerobic’ because oxygen is produced from nitrite in anaerobic conditions and is then used to aerobically oxidize methane (Ettwig et al, 2010). These theories are discussed in greater detail below. Research is still being conducted to determine whether just one theory or both theories explain the method(s) by which microbial nitrate oxidation of methane occurs.
Microbial Consortia Theory
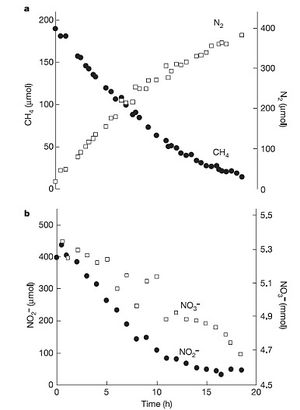
This theory hypothesizes that a microbial consortia consisting of an archaeon, Methanosarcinales, and bacteria, from the NC10 phylum, is required for the coupling of denitrification and anaerobic oxidation of methane (Raghoebarsing et al, 2006). Freshwater, anoxic sediment was collected from a canal in the Netherlands. The sediment was incubated and provided with carbon-13 labeled methane (electron donor), nitrite (electron acceptor), nitrate (electron acceptor), bicarbonate and essential trace elements. They found that nitrate, nitrite and methane were being consumed and that dinitrogen gas (N2) was being produced (Figure 1). Figure 1 shows that more nitrite was consumed by the culture than nitrate, indicating that nitrite is a better and therefore preferred electron acceptor. In addition, the denitrification rate was 290.1% faster with methane addition suggesting that denitrification and methane oxidation are coupled.
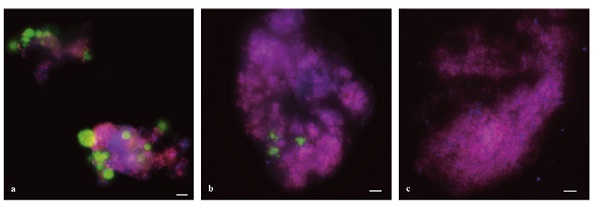
To ensure that both archaea and bacteria were participating in the coupling of denitrification and methane oxidation, Raghoebarsing et al (2006) measured the absorption of methane into the membrane lipids of the bacteria and archaea. To do this, they measured depletion of 13C in the membrane and compared it to that of methane before and after exposure to methane. They found that bacterial biomarkers were labeled more quickly and were more depleted in 13C than archaeal biomarkers; in fact only one archaeal biomarker showed signs of significant 13C depletion. The slow incorporation of methane into biomass by the archaeon suggests slow growth. Fluorescence in situ hybridization (FISH) analysis of culture biomass showed the presence of bacteria and archaea in the consortium. Archaea only comprised of 10% of the consortium, also indicating slow growth.
The mechanism by which the coupling occurs has not been determined, but is thought to occur in a manner similar to sulphate-dependent methane oxidation. It is proposed that the archaea oxidize methane to CO2 after which they transfer electrons to the bacteria which can then reduce nitrate/nitrite to N2 (Conrad, 2009). The coupling provides the bacteria and archaea with the thermodynamic conditions required for denitrification and methane oxidation, respectively. The following equations represent the overall reactions of methane with nitrate (NO3-) and nitrite (NO2-) (Raghoebarsing et al, 2006):
5CH4 + 8NO3- + 8H+ --> 5CO2 + 4N2 + 14H2O (G = -765 kJ mol-1 CH4)
3CH4 + 8NO2- + 8H+ --> 3CO2 + 4N2 + 10H2O (G = -928 kJ mol-1 CH4)
Intra-aerobic Denitrification Pathway Theory
Ettwig et al (2008) found that methane could be anaerobically oxidized in the absence of archaea. The researchers experimented with the same enriched culture, in anoxic conditions, used by Raghoebarsing and colleagues (2006) over a 22 month period. The culture was also provided with the same nutrients, electron donors and electron acceptors. They found that archaeal composition reduced over the time period and could not be detected by the fifteenth month using FISH analysis of biomass (Figure 2). Despite this decrease, the bacteria continued to oxidize methane. This suggests one of two things:
1.archaea and bacteria are both able to anaerobically oxidize methane completely separately, but the bacteria is able to outcompete the archaea or,
2.if archaea and bacteria are present, it is beneficial for them to form a syntrophic relationship and divide the labour; in an event where the archaea disappears, the bacteria can revert to an alternative pathway which allows it to continue with denitrification coupled to methane oxidation.
The bacterium responsible for this is a denitrifying methanotrophic bacterium in the NC10 phylum named Candidatus Methylomirabolis oxyfera, or M. oxyfera. It is a slow-growing, rod-shaped and gram negative bacterium. It was isolated by Ettwig and colleagues (2008, 2010). To date, it has only been found in freshwater sediments at mesophilic temperatures and pH.
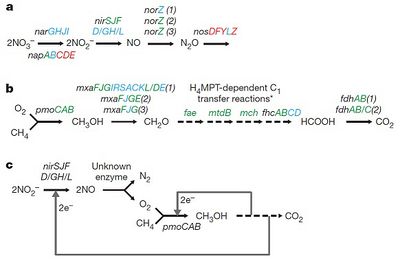
The metagenomes of two cultures of M. oxyfera from enriched freshwater sediments (collected from Australia and the Netherlands) were sequenced to piece together the mechanism of nitrite-dependent anaerobic oxidation of methane (Ettwig et al, 2010). The researchers found that M. oxyfera expressed genes for the conversion of NO3- to NO2-, NO2- to NO (nitric oxide) and NO to N2O (nitrous oxide), but no genes for the conversion of N2O to N2 (Figure 3a). Despite this, N2O could only be found in trace amounts. This suggests that the NO is being converted to N2 using an alternative mechanism where N2O is not an intermediate. On the other hand, it may be that N2O is being reduced to N2 using, not N2O reductase, but a different enzyme.
Ettwig et al (2010) also found that the metagenome did not contain genes coding fumarate-adding glycl-radical enzymes or methyl-coenzyme M reductase, either of which can be used for anaerobic oxidation of methane. Rather, they found that the metagenome coded for the aerobic oxidation of methane (Figure 3b). First, methane reacts with oxygen, using the membrane-bound form of the enzyme methane mono-oxygenase (pMMO), to form methanol (CH3OH) and water. The pMMO complex is thought to function either by aiding oxygen production and then consuming the oxygen produced or by using NO directly as an oxygen source. Further research needs to be conducted to establish the mechanism by which pMMO works in M. oxyfera.
A separate enzyme, methanol dehydrogenase, is used to form formaldehyde (CH2O) from CH3OH. CH2O, in turn, is oxidized to form formate (HCOO-). The conversion of CH2O to HCOO- requires a tetrahydromethanopterin-dependent C1 transfer module which is thought to be a primordial metabolic molecule; this indicates that M. oxyfera is from a particularly deep-branching lineage. Finally, HCOO- is converted to CO2 using the enzyme formate dehydrogenase. The electron flow resulting from the oxidation of methane generates a proton motive force and therefore allows for ATP synthesis (Ettwig et al, 2010).
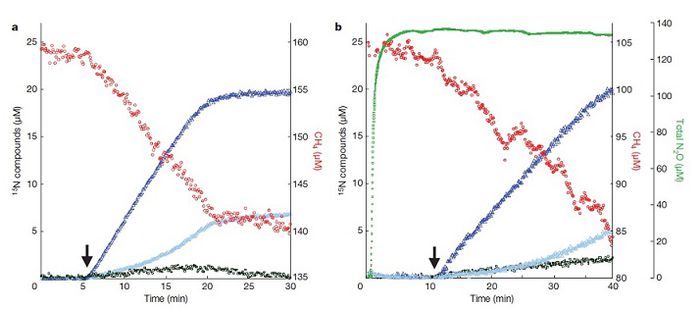
A peculiar aspect of the pathway is that nitrite is required for the mechanism to proceed even though M. oxyfera contains genes that code nitrate reductase (Figure 3a). Ettwig et al (2010) conducted an experiment where they provided an enrichment culture, under anoxic conditions, with nitrate and carbon-13 labeled CH4. Methane oxidation did not occur and N2 gas was not produced when the culture was incubated. However, as soon as nitrogen-15 labeled nitrite was added, methane oxidation commenced and labeled N2 gas was formed in stoichiometric quantities, indicating that N2 was produced solely from the added nitrite (Figure 4a). Is nitrate reductase not produced even though the metagenome codes for it?
Nevertheless, these findings suggest that incomplete denitrification somehow results in the production of oxygen and N2 gas (Figure 3c). Ettwig et al (2010) suggest that NO is converted to N2 and oxygen in a one-step mechanism using an ‘NO dismutase’ enzyme. The proposed NO dismutase enzyme, however, could not be isolated by the researchers. Further research needs to be conducted to isolate and identify the enzyme(s) responsible for the conversion of NO to N2 and oxygen.
Assuming that the aforementioned reaction occurs, how is the NO reductase enzyme (used to reduce NO to N2O) coded by the metagenome relevant in M. oxyfera's metabolic system? In other bacterial species, NO reductase has been used to detoxify NO as NO exposure can induce cell stress (Frey and Kallio, 2005). The prediction that N2O was not an intermediate for N2 production or a suitable electron acceptor for methane oxidation was tested by incubating the enrichment culture with 13CH4, nitrate and N2O (Ettwig et al, 2010). The researchers found that neither methane nor N2O were consumed, as per the prediction (Figure 4b). The role of NO reductase needs to be determined in future experiments.
Implications
Evolutionary Significance
The mechanism(s) of microbial nitrate oxidation of methane oxidation has evolutionary importance. It was, for example, originally thought that the three main oxygen-producing biological pathways were photosynthesis, chlorate respiration and the detoxification of reactive oxygen species and that these were the processes responsible for oxygenating the atmosphere (Ettwig et al, 2010). The intra-aerobic pathway, discovered by Ettwig et al (2010), may have also contributed to atmospheric oxygen levels because it is likely that some of the oxygen produced from NO escaped before it could be used for methane oxidation.
Moreover, the pathways proposed by Ettwig et al (2010) and Raghoebarsing et al (2006) may have evolved before photosynthesis and respiration. In the Archaen Eon, conditions were anaerobic and the atmosphere largely consisted of methane. In addition, Ducluzeau et al (2009) proposed that NO, nitrite and nitrate were widely available during the Archaen Eon as strong oxidants. Microorganisms using nitrate-dependent oxidation of methane could have thrived in such conditions. These pathways, therefore, also likely preceded aerobic respiration as aerobic respiration is thought to have evolved after photosynthesis (Ducluzeau et al, 2009). The intra-aerobic pathway, especially, may have actually allowed for the development of niches for the evolution of aerobic metabolism due to the local production of oxygen (Ettwig et al, 2010).
Global Climate Change
The global budget of methane in the atmosphere is 500-600 Tg CH4 per year. Its levels have increased between pre-industrial times and now from 715 ppbv to 1770 ppbv. Much of this increase can be attributed to fossil fuel burning and the intensification of agriculture – ruminants and large-scale rice plantation produce substantial amounts of methane (Conrad, 2009). The increase of methane in the atmosphere has significantly contributed to climate change, given its status as a greenhouse gas with a much higher global warming potential that carbon dioxide.
Climate warming has a positive feedback loop. In the case of wetlands, for example, warming has accelerated the actions of soil methanogens, thereby increasing the release of methane from the soil. Since methane is a potent greenhouse gas, the increase in methane levels will only increase warming. This, in turn will further increase methane release from the soil provided that soil moisture levels remain high (Cao et al, 1998).
Methane-oxidizing bacteria, as mentioned earlier, can mediate methane fluxes and nitrate levels. It may, therefore, be possible to use them for bioremediation. Nitrate/nitrite-dependent methane-oxidizing microbes can be used to offset increases in methane production (Raghoebarsing et al, 2006). In the case of agricultural run-off, the same microbes can be used to consume the nitrate/nitrite in soils and aquatic and marine sediments. These microbes can also be used to restore eutrophic marine and freshwater ecosystems as they thrive in anoxic conditions where there is a lot of nitrite and methane available. The use of the intra-aerobic pathway may help re-oxygenate the ecosystem, allowing the ecosystem to sustain life once again.
Methane Gas Hydrates
The rise in atmospheric temperature as a result of climate change has caused the oceans to warm. As the oceans continue to warm, methane hydrates become more and more unstable. Methane hydrates, or methane clathrates, are structures that can be found on the ocean floor. They are made up of water crystals and can trap enormous amounts of methane gas within the resulting lattice. It is estimated that these methane reservoirs hold between 500,000 and 1,000,000 Tg of methane (Conrad, 2009). Methane hydrates require low temperatures and low pressures or moderate temperatures and high pressures to remain stable (Biello, 2006).
At present, a large proportion of the methane they release (70-300 Tg CH4 per year) is consumed by methane-oxidizing microbes, meaning that methane hydrate emissions only compose about 1% of the total atmospheric methane budget (Conrad, 2009). The actions of methane-oxidizing microbes are, therefore, vital in mediating upward methane fluxes from oceans. However, if the oceans continue to warm, it may well be that there will be explosive releases of methane from the methane hydrates which cannot be contained by methane-oxidizing microbial species. This would prove to be catastrophic for all ecosystems and the life that they sustain.
Ettwig et al (2008, 2010) and Raghoebarsing et al (2006) isolated the enriched samples of nitrate/nitrite-dependent anaerobic methane oxidizers from freshwater systems. So far, none have been isolated from marine sediments. Part of this may be due to the fact that gas hydrates tend to be in parts of the ocean that are difficult and dangerous to access. It is possible that there are nitrate/nitrite-dependent anaerobic methane oxidizers in the deep ocean (Valentine, 2011). It would be interesting to study the ability of M. oxyfera to survive and function in deep ocean-like conditions.
Conclusion
Overall paper length should be 3,000 words, with at least 3 figures.
References
Oremland, RS. 2010. NO connection with methane. Nature 464: 500-501.
Edited by student of Joan Slonczewski for BIOL 238 Microbiology, 2009, Kenyon College.