Prochlorococcus marinus 2012: Difference between revisions
| Line 70: | Line 70: | ||
5. Liu, Hongbin, Hector A. Nolla, and Lisa Campbell. "Prochlorococcus Growth Rate and Contribution to Primary Production in the Equatorial and Subtropical North Pacific Ocean." Aquatic Microbial Ecology 12 (1997): 39-47. | 5. Liu, Hongbin, Hector A. Nolla, and Lisa Campbell. "Prochlorococcus Growth Rate and Contribution to Primary Production in the Equatorial and Subtropical North Pacific Ocean." Aquatic Microbial Ecology 12 (1997): 39-47. | ||
6. | **6. Tolonen, Andrew Carl. "Prochlorococcus genetic transformation and genomics of nitrogen metabolism". Woods Hole Oceanographic Institution. Massachusetts Institute of Technology. 2005. | ||
7. | 7. Chisholm, Sallie W., Sheila L. Frankel, Ralf Goericke et al. "Prochlorococcus marinus nov. gen. nov. sp.: an oxyphototrophic marine prokaryote containing divinyl chlorophyll a and b." Archives of Microbiology 157: 3 (1992): 297-300. | ||
8.Kolowrat, Christian; Partensky, Frédéric; Mella-Flores, Daniella; Corguillé, Gildas Le; Boutte, Christophe; Blot, Nicolas; Ratin, Morgane; Ferréol, Martial; Lecomte, Xavier; Gourvil, Priscillia; Lennon, Jean-François; Kehoe, David M.; Garczarek, Laurence. "Ultraviolet stress delays chromosome replication in light/dark synchronized cells of the marine cyanobacterium Prochlorococcus marinus." BMC Microbiology, 2010, Vol. 10, p204-227 | 8. Steglich, Caludia., Matthias Futschik, Trent Rectro, Robert Steen, and Sallie W. Chisholm. "Genome-Wide Analysis of Light Sensing ni Prochlorococcus." Journal of Bacteriology 188: 22. (Nov. 2006): 7796-7806. | ||
9.Kolowrat, Christian; Partensky, Frédéric; Mella-Flores, Daniella; Corguillé, Gildas Le; Boutte, Christophe; Blot, Nicolas; Ratin, Morgane; Ferréol, Martial; Lecomte, Xavier; Gourvil, Priscillia; Lennon, Jean-François; Kehoe, David M.; Garczarek, Laurence. "Ultraviolet stress delays chromosome replication in light/dark synchronized cells of the marine cyanobacterium Prochlorococcus marinus." BMC Microbiology, 2010, Vol. 10, p204-227 | |||
Revision as of 06:30, 17 February 2012
A Microbial Biorealm page on the genus Prochlorococcus marinus 2012 Prochlorococcus
Classification
Higher order taxa
Bacteria;Cyanobacteria; Prochlorales; Prochlorococcaceae; Prochlorococcus
|
NCBI: Taxonomy |
Species
|
NCBI: Genome |
Prochlorococcus marinus
Description and significance
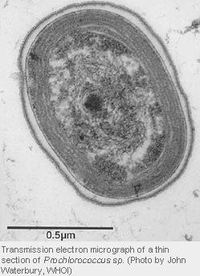
Prochlorococcus is a photosynthetic prokaryote. It is revealed by transmission election microscopy to have the same structure as a typical cyanacobacteria[3]. The cytoplasm holds DNA fibrils, carboxyzomes, and glycogen granules, which can be found near or between thylakoids. There is commonly two to four thylakoids in a Prochlorococcus cell, although sometimes there can be up to six thylakoids. Thylakoids can be found running parallel to the cell membrane [4].
Many methods used to find cell sizes tend to be biased towards tiny things, including Prochlorococcus cells. It is estimated that Prochlorococcus cells sizes vary from 0.5 to 0.8 μm in length, and 0.4 to 0.6 μm in width. A Prochlorococcus cell's size van vary depending on its environment. For example, its size was shown to increase by 0.45 to 0.75 μm between the surface and at a depth of 150 meters in the Sargasso Sea [4]. Studies show that Prochlorococcus can grow quite quickly in nutrient-poor habitats. The small size allows these picoplankton to take up dissolved nutrients more efficiently than larger organisms [5].
Prochlorococcus are important due to their contribution to oxygen production. Their production is found to contribute up to 39% of primary production in the eastern and western equatorial Pacific, and up to 82% in the North Pacific Ocean at Station ALOHA.
Genome structure
Because of its small size, Prochlorococcus marinus usually has a small genome1. It actually maintains the smallest genome of all cyanobacteria, with only 1.7 Mb [1]. This may be considered an adaptation because in the ocean (environment of this organism) there is not an excess nitrogen or phosphorus, which are essential in the production of nucleotides. There are generally two strains of this bacteria found in the ocean. The low-light adapted and high- light adapted strains are a result of the difference of light levels at different depths in the ocean. The low- light adapted strains have extra genes for the antenna complex protein, which is where the chlorophyll gets its energy from light [1]. These low-light adapted strains genetically produce chlorophyll b as opposed to chlorophyll a (high-light adapted strain) because chlorophyll b can be more helpful at a with a wider spectrum of light, which is essential when light is scarce. By having genes for more chlorophyll b and antenna protein complex, these bacteria are able to thrive even at depths of up to 200m [1]. These low-light adapted strains have a lot more variation when it comes to their genome, most likely as an adaptation for the tougher environment that the deeper ocean poses. The ocean is a very stable environment, thus these microbes have seen a decrease in the amount of stress- related and housekeeping genes, as compared to the genome of other cyanobacteria [1]. The genes for antenna protein are increased, as we discussed their characteristics that are essential to this bacteria. The genome of these bacteria are essential in the studying of photosynthetic bacteria, and their response to changing environments because of their unique and decreased genome [1].
Cell structure and metabolism
Interesting features of cell structure; how it gains energy; what important molecules it produces.
Prochlorococcus marinus has been established as one of the smallest living cyanobacteriumAll photosyntheic organisms must adapt to light levels in their environment in order to survive. Since it is know that prochlorococcus marinus are oxyphotobacteria that thrive in the ocean between 40 degrees north and 40 degrees south, their cellular strutres should reflect survival components of this environment. Indeed, survival in a oligotrophic environment requires adaptations such as low cellular nutrient requirement and highly efficient nutrient transport systems. The mechanisms that prochlorococcus uses to acquire and metabolize nitrogen, phosphorus, and other elements in its environment are central to its ecology [6].
For example, prochlorococcus marinus is a photosyntheitic organism, whos cellular strucutre lacks phycobiliproteins. Phycobiliproteins are water-soluable proteins that are present in most cyanobacteria and act to capture light energy which can then be used during photosynthesis. Instead of these proteins, prochlorococcus uses divinyl-chlorophyll a and b as their primary syntheitc pigments used during the photosyntheic process to gain energy [7]. This specialized pigment apparatus used to capture light is specific to prochlorococcus marinus which results in a unique light absorption spectrum, and sets it apart from other marine bacteria such as other plants and alge [8].
Ecology
Habitat; symbiosis; contributions to the environment.
Pathology
How does this organism cause disease? Human, animal, plant hosts? Virulence factors, as well as patient symptoms.
Current Research
Enter summaries of the most recent research here--at least three required
Cool Factor
Describe something you fing "cool" about this microbe.
References
1. Unknown Authors. "One of the Smallest Photosynthetic Organisms Known." Site Du Genoscope. Genoscope, 16 Jan. 2008. Web. 02 Feb. 2012. <http://www.cns.fr/spip/Prochlorococcus-marinus-smallest.html>.
3. Partensky, F., W. R. Hess, and D. Vaulot. "Prochlorococcus, a Marine Photosynthetic Prokaryote of Global Significance." Microbiology and Molecular Biology Reviews 63.1 (1999): 106-27.
4. Campbell, Lisa, H. A. Nolla, and Daniel Vaulot. "The Importance of Prochlorococcus to Community Structure in the Central North Pacific Ocean." Limnology and Oceanography 39.4 (1994): 954-61.
5. Liu, Hongbin, Hector A. Nolla, and Lisa Campbell. "Prochlorococcus Growth Rate and Contribution to Primary Production in the Equatorial and Subtropical North Pacific Ocean." Aquatic Microbial Ecology 12 (1997): 39-47.
- 6. Tolonen, Andrew Carl. "Prochlorococcus genetic transformation and genomics of nitrogen metabolism". Woods Hole Oceanographic Institution. Massachusetts Institute of Technology. 2005.
7. Chisholm, Sallie W., Sheila L. Frankel, Ralf Goericke et al. "Prochlorococcus marinus nov. gen. nov. sp.: an oxyphototrophic marine prokaryote containing divinyl chlorophyll a and b." Archives of Microbiology 157: 3 (1992): 297-300.
8. Steglich, Caludia., Matthias Futschik, Trent Rectro, Robert Steen, and Sallie W. Chisholm. "Genome-Wide Analysis of Light Sensing ni Prochlorococcus." Journal of Bacteriology 188: 22. (Nov. 2006): 7796-7806.
9.Kolowrat, Christian; Partensky, Frédéric; Mella-Flores, Daniella; Corguillé, Gildas Le; Boutte, Christophe; Blot, Nicolas; Ratin, Morgane; Ferréol, Martial; Lecomte, Xavier; Gourvil, Priscillia; Lennon, Jean-François; Kehoe, David M.; Garczarek, Laurence. "Ultraviolet stress delays chromosome replication in light/dark synchronized cells of the marine cyanobacterium Prochlorococcus marinus." BMC Microbiology, 2010, Vol. 10, p204-227