Desulforudis audaxviator: Difference between revisions
No edit summary |
No edit summary |
||
| Line 3: | Line 3: | ||
=Classification= | =Classification= | ||
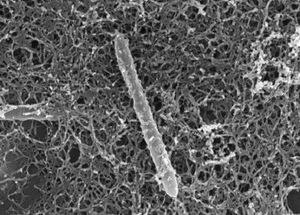
[[File:D._audaxviator.jpg|300px|thumb|right|''D. audaxviator'']] | |||
Kingdom: Bacteria | Kingdom: Bacteria | ||
| Line 58: | Line 58: | ||
==Energy== | ==Energy== | ||
It is thought that a long-term seclusion from O₂ has lead to the complete loss of a system for oxygen tolerance, making ''D. audaxviator'' an obligate anaerobe.[[#References|[4]]][[#References|[8]]] Since the bacterium cannot utilize O₂ as its electron acceptor, it uses sulfate (SO₄²¯), an energetically favorable alternative to O₂.[[#References|[4]]] However, it is generally sulfide (H₂S) that is naturally available from the surrounding rock. Radioactivity generated by the decay of Uranium in the rocks splits ambient water molecules into H⁺ and hydroxide (OH), which then forms hydrogen peroxide (H₂O₂). The hydrogen peroxide reacts with the environmental sulfide to form sulfate. The bacteria can then use the sulfate for dissimulatory sulfate reduction, returning the compound to sulfide. Electrons from the hydrogen left over from the splitting of water are used by the cell to reduce sulfate.[[#References|[7]]][[#References|[8]]] | It is thought that a long-term seclusion from O₂ has lead to the complete loss of a system for oxygen tolerance, making ''D. audaxviator'' an obligate anaerobe.[[#References|[4]]][[#References|[8]]] Since the bacterium cannot utilize O₂ as its electron acceptor, it uses sulfate (SO₄²¯), an energetically favorable alternative to O₂.[[#References|[4]]] However, it is generally sulfide (H₂S) that is naturally available from the surrounding rock. Radioactivity generated by the decay of Uranium in the rocks splits ambient water molecules into H⁺ and hydroxide (OH), which then forms hydrogen peroxide (H₂O₂). The hydrogen peroxide reacts with the environmental sulfide to form sulfate. The bacteria can then use the sulfate for dissimulatory sulfate reduction, returning the compound to sulfide. Electrons from the hydrogen left over from the splitting of water are used by the cell to reduce sulfate.[[#References|[7]]][[#References|[8]]] | ||
==Nitrogen== | ==Nitrogen== | ||
Revision as of 18:54, 14 December 2012
Classification
Kingdom: Bacteria
Phylum: Firmicutes
Class: Clostridia
Order: Clostridiaceae
Family: Peptococcaceae
Genus: Candidatus Desulforudis
Species: Candidatus Desulforudis audaxviator
The name “audaxviator” was inspired by a quote from the book Journey to the Center of the Earth, by Jules Verne. The Latin phrase “audax viator” translates to “bold traveler”, a reference to the bacterium’s subterranean habitat.[4][8]
Description and Significance
D. audaxviator was first discovered by NAI’s Indiana-Princeton-Tennessee Astrobiology Initiative’s (IPTAI) team members in 2005/2006.[8] It was found in a South American gold mine, at a depth that was previously thought to be uninhabited by life.[3] D. audaxviator was found at level 104, 2.8km deep in the Mponeng gold mine.[4] The groundwater in which they were found exceeded 60° C, had a pH of 9.3, was anoxic, and was severely nutrient deficient.[4][7]
D. audaxviator is a rod shaped Gram-positive bacterium. They are motile, sporulating, sulfate reducing chemoautotrophs, and are categorized as thermophiles and obligate anaerobes.[4][6] These bacteria are the only known resident of their environment, and therefore live independently of other organisms.[4][8] This discovery revealed that it is possible for a single genome to encode all proteins necessary for the biological portion of a basic ecosystem.[4][8] D. audaxviator comprises the only known ecosystem relies on radioactivity instead of sunlight, or chemical energy from the earth (see section on metabolism).[7][8]
D. audaxviator and other deep subterranean microorganisms have been heavily researched for several reasons. D. audaxviator’s simplistic environment coupled with its complete life-supporting genome makes it a good representation of an organism that could be capable of life on an alien planet.[7][8] This bacterium performs all of the processes needed to sustain life if given a source of radioactivity, sulfate, water, ammonia and carbon dioxide, all of which are thought to be present on rocky planets such as Mars.[8] D. audaxviator could also be a model of prehistoric life on Earth, providing valuable insight to the origins of life. [3][7]
Genome Structure
D. audaxviator’s genome is 2.35 megabase pairs (Mbp) in size, containing 2157 protein encoding genes.[4] This is larger than the expected <2000 genes found in other free-living organisms.[4]
Other details of the genome[4]:
- 86.8% of the genome codes for protein
- The G+C content of the genome is 60.9%
- The average length of an open reading frame is 910 base pairs
Horizontal Gene Transfer
Certain genes in the D. audaxviator genome are considered to be products of archaeal horizontal gene transfer.[4] Some of these genes and gene products include an extra copy of an archaeal-type sulfate adenylyltransferase (to convert ATP and sulfate to diphosphate and adenylyl sulfate), a H⁺ translocating pyrophosphatase (to convert pyrophosphate into two phosphate ions), a nitrogenase (to fix N₂ gas), CRISPR-associated genes (useful in viral defense), a 2nd carbon monoxide dehydrogenase system (to convert CO into CO₂), and genes for formation of gas vesicles.[4]
The archaeal genes present in the D. audaxviator genome could imply that it or an ancestor was once in contact with an archaeal species.[10] For example, D. audaxviator possesses a carbon dioxide fixing pathway that is similar to that of archaea.[3] As evidence that this could have happened, Takai et al. (2001) researched life in other mine shafts, revealing that archaea can live at extreme depths. Samples were taken from the East and West Driefontein gold mines, the Kloof mines, and the Beatrix Mine. All three of these mines are located in the Witswatersrand basin, the same basin as the Mponeng gold mine. Archaeal species were found in fissure water 0.87km to 3.08km deep. These species had high C+G content (57-60%) like D. audaxviator, although most were determined to be methanogens.[9]
Related Bacterial Species
Similar bacteria have been found in other environments and locations. Alawi et al. (2011) found a related bacterium (94% genome similarity) in wells 3.5km deep under a geothermal plant in South Germany.[1] A different study by Balci et al. (2012) revealed another D. audaxviator-like bacterium (94% genome similarity) to be living in Lake Maslak sediments in Istanbul, Turkey. This bacterium, although not living at a great depth, is also a sulfate reducing bacterium.[2] Another bacterium (91% genome similarity) was detected by Itävaara et al. (2011), it was found between 1.2 and 1.5km deep in the Outokumpu borehole, drilled in Eastern Finland.[5]
Metabolism
D. Audaxviator is a sulfate reducing chemoautotroph, capable of fixing nitrogen and carbon.[4] It is capable of forming endospores when the environmental conditions become too unfavorable.[6] D. audaxviator has the cellular machinery to sense nutrients and is flagellated, allowing the bacteria to perform chemotaxis.[4] The cell also has every pathway needed to synthesize all amino acids.[4]
Energy
It is thought that a long-term seclusion from O₂ has lead to the complete loss of a system for oxygen tolerance, making D. audaxviator an obligate anaerobe.[4][8] Since the bacterium cannot utilize O₂ as its electron acceptor, it uses sulfate (SO₄²¯), an energetically favorable alternative to O₂.[4] However, it is generally sulfide (H₂S) that is naturally available from the surrounding rock. Radioactivity generated by the decay of Uranium in the rocks splits ambient water molecules into H⁺ and hydroxide (OH), which then forms hydrogen peroxide (H₂O₂). The hydrogen peroxide reacts with the environmental sulfide to form sulfate. The bacteria can then use the sulfate for dissimulatory sulfate reduction, returning the compound to sulfide. Electrons from the hydrogen left over from the splitting of water are used by the cell to reduce sulfate.[7][8]
Nitrogen
D. audaxviator has an ammonium (NH₄⁺) transporter that takes up ammonia (NH₃) from the surrounding environment. Ammonia is thought to be at a high enough concentration to provide a sufficient source of nitrogen.⁽⁴⁾ The cell has glutamine synthetases to derive nitrogen from ammonia, but it also has a temperature resistant nitrogenase.[4] The nitrogenase allows D. audaxviator to use the more energetically expensive method of obtaining nitrogen from N₂, if ammonia is not available.[4]
Carbon
D. audaxviator obtains carbon from many sources. The bacterium has transporters for both sugars and amino acids, which can be used a carbon source if they are available in the environment.[4] This may occur when another bacterium dies.⁽⁴⁾ D. audaxviator can also obtain carbon from inorganic sources. A putative carbonate adenosine triphosphate (ATP) binding cassette transporter or a putative bicarbonate/Na⁺ symporter is responsible for the uptake of CO₂.[4] A carbon monoxide dehydrogenase is used in the CoA synthesis pathway to assimilate this inorganic carbon. Formate and CO may also be used as a carbon source.[4]
Ecology
D. audaxviator lives in an ecosystem by itself, so it does not naturally interact with any other organisms.[4] The bacterium interacts with its environment on its hunt to find nutrients. The bacterium performs chemotaxis along chemical gradients, and takes up nutrients it needs. This may include taking up nutrients recycled from dead cells.[4]
References
(1) Alawi, M., Lerm, S., Vetter, A., Wolfgramm, M., Seibt, A., and Würdemann, H. “Diversity of Sulfate-Reducing Bacteria in a Plant Using Deep Geothermal Energy.” Grundwasser, 2011, DOI: 10.1007/s00767-011-0164-y
(2) Balci, N., Vardar, N., Yelboga, E., and Karaguler, N. “Bacterial Community Composition of Sediments from Artificial Lake Maslak, Istanbul, Turkey.” Environmental Monitoring and Assessment, 2012, DOI: 10.1007/s10661-011-2368-0
(3) Bonch-Osmolovskaya, E. “High-Temperature Deep-Subsurface Microbial Communities as a Possible Equivalent of Ancient Ecosystems.” Paleontological Journal, 2010, DOI: 10.1134/S0031030110070130
(4) Chivian,D., Brodie, E., Alm, E., Culley, D., Dehal, P., DeSantis,T., Gihring, T., Lapidus, A., Lin, L., Lowry, S., Moser, D., Richardson, P., Southam, G., Wanger, G., Pratt, L., Andersen, G., Hazen, T., Brockman, F., Arkin, A., and Onstott, T. “Environmental Genomics Reveals a Single-Species Ecosystem Deep Within Earth.” Science, 2008, DOI: 10.1126/science.1155495
(5) Itävaara, M., Nyyssönen, M., Kapanen, A., Nousiainen, A., Ahonen, L., and Kukkonen, I. “Characterization of Bacterial Diversity to a Depth of 1500m in the Outokumpu Deep Borehole, Fennoscandian Shield.” FEMS Microbiology Ecology, 2011, DOI: 10.1111/j.1574-6941.2011.01111.x
(6) Lin, L., Wang, P., Rumble, D., Lippmann-Pipke, J., Boice, E., Pratt, L., Sherwood Lollar, B., Brodie, E., Hazen, T., Andersen, G., DeSantis, T., Moser, D., Kershaw, D., and Onstott, T. “Long-Term Sustainability of a High-Energy, Low-Diversity Crustal Biome.” Science, 2006, DOI: 10.1126/science.1127376
(7) Mascarelli, A. “Geomicrobiology: Low Life.” Nature, 2009, DOI: 10.1038/459770a
(8) Scalice, D. “Life Without the Sun.” NASA Astrobiology, 2008, LINK: https://astrobiology.nasa.gov/articles/2008/10/10/life-without-the-sun/
(9) Takai, K., Moser, D., DeFlaun, M., Onstott, T., and Fredrickson, J. “Archaeal Diversity in Waters from Deep South African Gold Mines.” Applied and Environmental Microbiology, 2001, DOI: 10.1128/AEM.67.21.5750-5760.2001
(10) Thomas, C., and Nielsen, K. “Mechanisms of, and Barriers to, Horizontal Gene Transfer between Bacteria.” Nature Reviews Microbiology, 2005, DOI: 10.1038/nrmicro1234