Lechuguilla cave microbiome: Difference between revisions
No edit summary |
No edit summary |
||
| Line 10: | Line 10: | ||
Lechuguilla Cave is an extreme environment defined by darkness and limited nutrients. Nonetheless, diverse microbial communities inhabit the aphotic and oligotrophic surfaces of the cave in rich biofilms. These communities are anchored by chemolithoautotrophic primary producers who oxidize ferromanganese substances in the rock, leaving distinctive deposits of ferromanganese ores nearby [[#References|[5]]]. These chemolithoautotrophs support secondary populations of chemoheterotrophic bacteria, archaea, and fungi [[#References|[6]]]. | Lechuguilla Cave is an extreme environment defined by darkness and limited nutrients. Nonetheless, diverse microbial communities inhabit the aphotic and oligotrophic surfaces of the cave in rich biofilms. These communities are anchored by chemolithoautotrophic primary producers who oxidize ferromanganese substances in the rock, leaving distinctive deposits of ferromanganese ores nearby [[#References|[5]]]. These chemolithoautotrophs support secondary populations of chemoheterotrophic bacteria, archaea, and fungi [[#References|[6]]]. | ||
[[File:Lechuguilla_cave_microbiome_figure_1.jpeg| | [[File:Lechuguilla_cave_microbiome_figure_1.jpeg|400px|thumb|left|Figure 1: Red-brown and brown ferromanganese deposits from survey site in Lechuguilla Cave [[#References|[7]]]. Close-up shows scale (cm) and texture of deposits]] | ||
Several fungal and bacterial phyla have been isolated from the cave. These include the fungi [https://microbewiki.kenyon.edu/index.php/Penicillium Penicillium], [https://microbewiki.kenyon.edu/index.php/Aspergillus Aspergillus], [https://microbewiki.kenyon.edu/index.php/Cylindrocladium Cylindrocladium], [https://microbewiki.kenyon.edu/index.php/Rhizopus Rhizopus], [https://microbewiki.kenyon.edu/index.php/Mycelia Mycelia sterilia], [https://microbewiki.kenyon.edu/index.php/Mucor Mucor], [https://microbewiki.kenyon.edu/index.php/Paecileomyces Paecileomyces], [https://microbewiki.kenyon.edu/index.php/Fusarium Fusarium], and [https://microbewiki.kenyon.edu/index.php/Epicoccum Epicoccum]; and the bacteria [https://microbewiki.kenyon.edu/index.php/Bacillus Bacillus], [https://microbewiki.kenyon.edu/index.php/Actinomycetes Actinomycetes], [https://microbewiki.kenyon.edu/index.php/Arthrobacter Arthrobacter], [https://microbewiki.kenyon.edu/index.php/Chryseomonas Chryseomonas luteola], [https://microbewiki.kenyon.edu/index.php/Rhodococcus Rhodococcus], and [https://microbewiki.kenyon.edu/index.php/Staphylococcus Staphylococcus] [[#References|[8]]]. | Several fungal and bacterial phyla have been isolated from the cave. These include the fungi [https://microbewiki.kenyon.edu/index.php/Penicillium Penicillium], [https://microbewiki.kenyon.edu/index.php/Aspergillus Aspergillus], [https://microbewiki.kenyon.edu/index.php/Cylindrocladium Cylindrocladium], [https://microbewiki.kenyon.edu/index.php/Rhizopus Rhizopus], [https://microbewiki.kenyon.edu/index.php/Mycelia Mycelia sterilia], [https://microbewiki.kenyon.edu/index.php/Mucor Mucor], [https://microbewiki.kenyon.edu/index.php/Paecileomyces Paecileomyces], [https://microbewiki.kenyon.edu/index.php/Fusarium Fusarium], and [https://microbewiki.kenyon.edu/index.php/Epicoccum Epicoccum]; and the bacteria [https://microbewiki.kenyon.edu/index.php/Bacillus Bacillus], [https://microbewiki.kenyon.edu/index.php/Actinomycetes Actinomycetes], [https://microbewiki.kenyon.edu/index.php/Arthrobacter Arthrobacter], [https://microbewiki.kenyon.edu/index.php/Chryseomonas Chryseomonas luteola], [https://microbewiki.kenyon.edu/index.php/Rhodococcus Rhodococcus], and [https://microbewiki.kenyon.edu/index.php/Staphylococcus Staphylococcus] [[#References|[8]]]. | ||
| Line 26: | Line 26: | ||
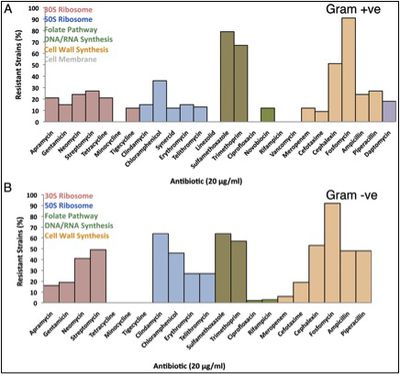
Given the importance of these metabolities in extreme environments, and given the colonization of Lechuguilla cave by members of the phyla that have provided humanity with its most common clinical antibiotic drugs, it is not surprisingly that isolates from the cave displayed widespread resistance to clinical antibiotics despite several million years of isolation in a pristine environment. Researchers from McMaster University, Canada and the University of Akron, USA isolated 93 strains from Lechuguilla Cave (33% Gram-positive and 63% Gram-negative) and tested them against 26 natural, semi-synthetic, and synthetic clinical antibiotics. Resistance to 3-4 different classes of antibiotic was observed in 70% of the Gram-positive isolates and 65% of the Gram-negative isolates [[#References|[17]]]. | Given the importance of these metabolities in extreme environments, and given the colonization of Lechuguilla cave by members of the phyla that have provided humanity with its most common clinical antibiotic drugs, it is not surprisingly that isolates from the cave displayed widespread resistance to clinical antibiotics despite several million years of isolation in a pristine environment. Researchers from McMaster University, Canada and the University of Akron, USA isolated 93 strains from Lechuguilla Cave (33% Gram-positive and 63% Gram-negative) and tested them against 26 natural, semi-synthetic, and synthetic clinical antibiotics. Resistance to 3-4 different classes of antibiotic was observed in 70% of the Gram-positive isolates and 65% of the Gram-negative isolates [[#References|[17]]]. | ||
[[File:Lechuguilla_cave_microbiome_figure_2.jpeg| | [[File:Lechuguilla_cave_microbiome_figure_2.jpeg|400px|thumb|left|Figure 2: Resistance levels of Lechuguilla Cave bacteria at 20 µg/ml against various antibiotics [[#References|[18]]]. Antibiotics are grouped by colour according to target of antibiotic action.]] | ||
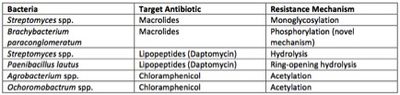
Additionally, they observed that antibiotics were inactivated using mechanisms very similar to those observed in surface bacteria. They also observed a novel inactivation mechanism that has thus far not been observed among surface microbes [[#References|[19]]]. | Additionally, they observed that antibiotics were inactivated using mechanisms very similar to those observed in surface bacteria. They also observed a novel inactivation mechanism that has thus far not been observed among surface microbes [[#References|[19]]]. | ||
[[File:Lechuguilla_cave_microbiome_figure_3.jpeg| | [[File:Lechuguilla_cave_microbiome_figure_3.jpeg|400px|thumb|right|Figure 3: Major resistance mechanisms observed in Lechuguilla Cave isolates.]] | ||
==Implications of Antibiotic Resistance in Lechuguilla Cave== | ==Implications of Antibiotic Resistance in Lechuguilla Cave== | ||
Revision as of 04:33, 27 December 2012
Introduction
Lechuguilla Cave is a deep and expansive cave in New Mexico, USA. It has roughly 216 km of passageway, and it is 489 m below the surface at its deepest point. The cave and its microbiome have attracted interest from microbiologists because, despite several million years of isolation from the terrestrial environment, bacterial isolates from the cave display widespread resistance to modern clinical antibiotic drugs. This discovery sheds new light on the origin of antibiotic resistance genes, and it offers new opportunities for developing effective next-generation antibiotic drugs.
Geology
Lechuguilla Cave is located in Carlsbad Caverns National Park, New Mexico. It formed between four and seven million years ago through hypogenic speleogenesis [1]. Hydrogen sulfide from hydrocarbons in the nearby Delaware Basin became oxidized to sulfuric acid, and acidified groundwater infiltrated upward into the carbonate rock of the Capitan Formation, thereby forming Lechuguilla Cave [2].
After formation, the aquifer dropped leaving the cave hydrologically isolated from phreatic groundwater. The cave is also hydrologically isolated from above by the impermeable rock of the Yates Formation that overlies the Capitan Formation [3]. As a result, the influx of terrestrial water occurs very slowly and in negligible quantities. The cave and its biota have remained effectively isolated from the terrestrial environment and human activity. Water infiltrating into the cave today would have left the surface long before the advent of human antibiotic use in agriculture and medicine [4].
Cave Environment and Biodiversity
Lechuguilla Cave is an extreme environment defined by darkness and limited nutrients. Nonetheless, diverse microbial communities inhabit the aphotic and oligotrophic surfaces of the cave in rich biofilms. These communities are anchored by chemolithoautotrophic primary producers who oxidize ferromanganese substances in the rock, leaving distinctive deposits of ferromanganese ores nearby [5]. These chemolithoautotrophs support secondary populations of chemoheterotrophic bacteria, archaea, and fungi [6].

Several fungal and bacterial phyla have been isolated from the cave. These include the fungi Penicillium, Aspergillus, Cylindrocladium, Rhizopus, Mycelia sterilia, Mucor, Paecileomyces, Fusarium, and Epicoccum; and the bacteria Bacillus, Actinomycetes, Arthrobacter, Chryseomonas luteola, Rhodococcus, and Staphylococcus [8].
Microbial Ecology
The extreme oligotrophic nature of the cave environment means that interactions within these microbial communities are defined by fierce competition over scarce nutrients and complex cooperation to make the most of whatever is consumed [9]. Both processes of competition and cooperation are mediated by secondary metabolite products, specifically those with antibiotic properties [10]. The antagonistic use of these antibiotic metabolites has been well researched. Many bacteria and fungi are known to secrete metabolities with antimicrobial activity to inhibit growth by competitors [11]. Indeed, almost all clinical antibiotic drugs are derived from natural metabolites, many of which are produced by surface relatives of the phyla found in Lechuguilla Cave [12].
But new research suggests that these metabolites are not only used for competitive purposes; they have also been implicated in the cooperative process of biofilm formation as signaling molecules [13]. Researchers at the University of British Columbia, Canada have demonstrated that these metabolites are often present in the environment in concentrations far below their minimum inhibitory concentration, suggesting additional functions beyond antimicrobial competition [14]. This research also found that growth in the presence of low concentrations of these metabolites can significantly alter transcriptional activity, suggesting that at low concentrations these metabolites act as signaling molecules [15].
The extensive use of these metabolites to mediate biofilm formation is not surprising in an extreme environment like Lechuguilla Cave. Biofilms engender cooperation within the microbial community. The exopolysaccharide matrix of the biofilm inhibits the diffusion of toxic compounds from the surroundings, facilitates the absorption of nutrients, protects the community from desiccation and other environmental factors, and promotes the exchange of genetic material so that adaptation to the habitat can be accelerated [16]. Biofilms confer a significant survival advantage to microbial communities living in extreme conditions.
Resistance of Lechuguilla Isolates
Whether used competitively as antimicrobial weapons or cooperatively as signaling molecules in biofilm communities, metabolites with antibiotic properties will be prevalent in harsh environments like Lechuguilla Cave. An important corollary is that genes encoding protection from the antimicrobial properties of these metabolites will be equally abundant. Microbial inhabitants will have to protect themselves from the metabolites of competitors and from the signaling molecules of their own microconsortia should they accumulate to dangerous concentrations.
Given the importance of these metabolities in extreme environments, and given the colonization of Lechuguilla cave by members of the phyla that have provided humanity with its most common clinical antibiotic drugs, it is not surprisingly that isolates from the cave displayed widespread resistance to clinical antibiotics despite several million years of isolation in a pristine environment. Researchers from McMaster University, Canada and the University of Akron, USA isolated 93 strains from Lechuguilla Cave (33% Gram-positive and 63% Gram-negative) and tested them against 26 natural, semi-synthetic, and synthetic clinical antibiotics. Resistance to 3-4 different classes of antibiotic was observed in 70% of the Gram-positive isolates and 65% of the Gram-negative isolates [17].
Additionally, they observed that antibiotics were inactivated using mechanisms very similar to those observed in surface bacteria. They also observed a novel inactivation mechanism that has thus far not been observed among surface microbes [19].
Implications of Antibiotic Resistance in Lechuguilla Cave
The prevalence of resistance found in the Lechuguilla Cave isolates along with the similarity in resistance mechanisms to those observed among terrestrial bacteria advances debate and inquiry into the origin of antibiotic resistance. The importance of human activity in the evolution resistance is controversial. There is consensus that medical and agricultural overuse has selected for and driven the emergence of resistant strains, but the origin of resistance itself and the role of environmental microbial ecologies as reservoirs of resistance is unclear.
Some research conducted on historical bacterial samples from the pre-antibiotic era showed an absence of resistance genes, suggesting that these are recently adaptations arising from human antibiotic activity [20]. Other research compared historical soil samples from the pre-antibiotic era with current soil samples. They found genetic indications of resistance before the advent of human antibiotic use, but observed a marked increase in the prevalence of resistance in the bacterial pangenome alongside the advent of human antibiotic use [21]. Both of these studies support the hypothesis that widespread antibiotic resistance is a recent phenomenon that developed directly in response to human activity.
The findings from Lechuguilla Cave support the opposing hypothesis: that widespread antibiotic resistance is an ancient, human-independent phenomenon. This corroborates other studies on microbes from other extreme and isolated habitats that also display antibiotic resistance. These studies suggest that a genetically rich resistome exists that is hard-wired into the bacterial pangenome.
Extreme isolated habitats provide useful opportunities to study the resistome, and the results of these studies have important clinical applications. By expanding our knowledge of the bacterial resistome, environments like Lechuguilla Cave can contribute to the development of an early warning system of resistance during the design of next-generation antibiotic drugs. These microbes could indicate whether some kind of resistance mechanism to a new clinical compound already resides in the resistome, presaging the rapid emergence of resistance to (and consequent ineffectiveness of) drugs under development [22].
Additionally, Lechuguilla Cave and other similar extreme habitats could provide fertile grounds in the search for new antibiotic templates. The depth and diversity of the resistome indicated by resistance findings at sites like Lechuguilla Cave suggest that countless metabolites with antibiotic properties remain to be discovered [23]. The intensity of competition and the complexity of cooperation in these oligotrophic habitats places great importance on these kinds of metabolites, making these kinds of extreme environments promising places to search for new clinical antibiotics.
References
1. Polyak V, Provencio PP. (2000) Summary of the Timing of Sulfuric-Acid Speleogenesis for Guadalupe Caves Based on Ages of Alunite. Journal of Cave and Karst Studies 62(2):72 – 74.
2. DuChene, HR. (2000) Bedrock Features of Lechuguilla Cave, Guadalupe Mountains, New Mexico. Journal of Cave and Karst Studies 62(2): 109 – 119.
3. Ibid.
4. Bhullar K, Waglechner N, Pawlowski A, Koteva K, Banks ED, Johnston MD, Barton HA, Wright GD. (2012) Antibiotic Resistance is Prevalent in an Isolated Cave Microbiome. PLoS ONE 7(4)
5. Northup DE, Barns SM, Yu LE, Spilde MN, Schelble RT, Dano KE, Crossey LJ, Connolly CA, Boston PJ, Natvig DO, Dahm CN. (2003) Diverse Microbial Communities Inhabiting Ferromanganese Deposits in Lechuguilla and Spider Caves. Environmental Microbiology 5(11): 1071 – 1086.
6. Cunningham KI, Northup DE, Pollastro RM, Wright WG, LaRock EJ. (1995) Bacteria, Fungi, and Biokarst in Lechuguilla Cave, Carlsbad Caverns National Park, New Mexico. Environmental Geology 25: 2 – 8.
7. Northup DE, Barns SM, Yu LE, Spilde MN, Schelble RT, Dano KE, Crossey LJ, Connolly CA, Boston PJ, Natvig DO, Dahm CN. (2003) Diverse Microbial Communities Inhabiting Ferromanganese Deposits in Lechuguilla and Spider Caves. Environmental Microbiology 5(11): 1071 – 1086.
8. Cunningham KI, Northup DE, Pollastro RM, Wright WG, LaRock EJ. (1995) Bacteria, Fungi, and Biokarst in Lechuguilla Cave, Carlsbad Caverns National Park, New Mexico. Environmental Geology 25: 2 – 8.
9. Gabriel CR, Northup DE. (2013) Microbial Ecology: Caves as an Extreme Habitat . In: Cheeptham N, ed. Cave Microbiomes: A Novel Resource for Drug Discovery. SpringerBriefs in Microbiology 1: 85 – 108.
10. De Lurdes M, Dapkevicius NE. (2013) Cave Biofilms and Their Potential for Novel Antibiotic Drug Discovery. In: Cheeptham N, ed. Cave Microbiomes: A Novel Resource for Drug Discovery. SpringerBriefs in Microbiology 1: 85 – 108.
11. Ibid.
12. Bhullar K, Waglechner N, Pawlowski A, Koteva K, Banks ED, Johnston MD, Barton HA, Wright GD. (2012) Antibiotic Resistance is Prevalent in an Isolated Cave Microbiome. PLoS ONE 7(4)
13. Davies J. (2006) Are antibiotics naturally antibiotics? Journal of Industrial Microbiology and Biotechnology 33: 496 – 499.
14. Ibid.
15. Ibid.
16. De Lurdes M, Dapkevicius NE. (2013) Cave Biofilms and Their Potential for Novel Antibiotic Drug Discovery. In: Cheeptham N, ed. Cave Microbiomes: A Novel Resource for Drug Discovery. SpringerBriefs in Microbiology 1: 85 – 108.
17. Bhullar K, Waglechner N, Pawlowski A, Koteva K, Banks ED, Johnston MD, Barton HA, Wright GD. (2012) Antibiotic Resistance is Prevalent in an Isolated Cave Microbiome. PLoS ONE 7(4)
18. Ibid.
19. Ibid.
20. Hughes VM, Datta N. (1983) Conjugative Plasmids in Bacteria of the ‘Pre-Antibiotic’ Era. Nature 302: 725 – 726.
21. Knapp CW, Dolfing J, Ehlert PA, Graham DW. (2010) Evidence of increasing antibiotic resistance gene abundance in archived soils since 1940. Environmental Science and Technology 44: 580 – 587.
22. Bhullar K, Waglechner N, Pawlowski A, Koteva K, Banks ED, Johnston MD, Barton HA, Wright GD. (2012) Antibiotic Resistance is Prevalent in an Isolated Cave Microbiome. PLoS ONE 7(4)
23. Ibid.