HIV-Derived Lentiviral Vectors and Their Use as Gene Therapy Agents Against Human Immunodeficiency Virus: Difference between revisions
| Line 18: | Line 18: | ||
<br><b>Brief Overview of the Life Cycle of HIV-1:</b><br> | <br><b>Brief Overview of the Life Cycle of HIV-1:</b><br> | ||
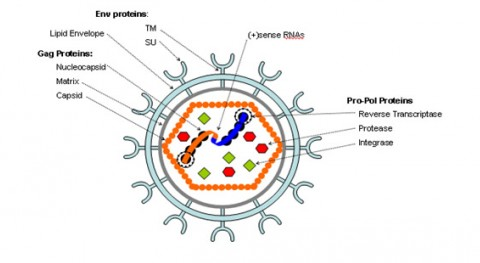
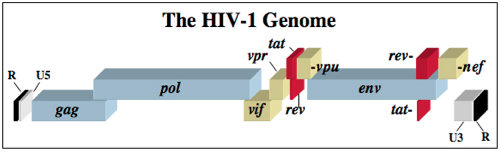
The HIV-1 virus consists of a diploid single-stranded RNA genome surrounded by a protein coat and outer phospholipid envelope (figure 1, [10]). Its genome contains three essential retroviral genes as well as well as regulatory genes that modulate host interactions (figure 2). The gag gene is responsible for glycoprotein structure, the env gene transcribes envelope proteins, and the pol gene transcribes the reverse transcriptase, responsible for producing a DNA copy of the HIV RNA genome [8]. Viral genes vif, vpr, vpu, rev, nef and tat transcribe virulence proteins [10]. All proviral genes, those that become integrated into the host (gag, pol, pro, vif, vpr, vpu, tat, and env), are bordered by two long terminal repeats (LTRs) essential for gene regulation [10]. Viral promoters and enhancers are located in the LTR U3 region [10]. The packaging signal, Ψ, is located in the 5’ end of the genome, and is responsible for packaging genes downstream of itself into the viral capsid during replication [10]. <br> | The HIV-1 virus consists of a diploid single-stranded RNA genome surrounded by a protein coat and outer phospholipid envelope (figure 1, [10]). Its genome contains three essential retroviral genes as well as well as regulatory genes that modulate host interactions (figure 2). The gag gene is responsible for glycoprotein structure, the env gene transcribes envelope proteins, and the pol gene transcribes the reverse transcriptase, responsible for producing a DNA copy of the HIV RNA genome [8]. Viral genes vif, vpr, vpu, rev, nef and tat transcribe virulence proteins [10]. All proviral genes, those that become integrated into the host (gag, pol, pro, vif, vpr, vpu, tat, and env), are bordered by two long terminal repeats (LTRs) essential for gene regulation [10]. Viral promoters and enhancers are located in the LTR U3 region [10]. The packaging signal, Ψ, is located in the 5’ end of the genome, and is responsible for packaging genes downstream of itself into the viral capsid during replication [10]. <br><br> | ||
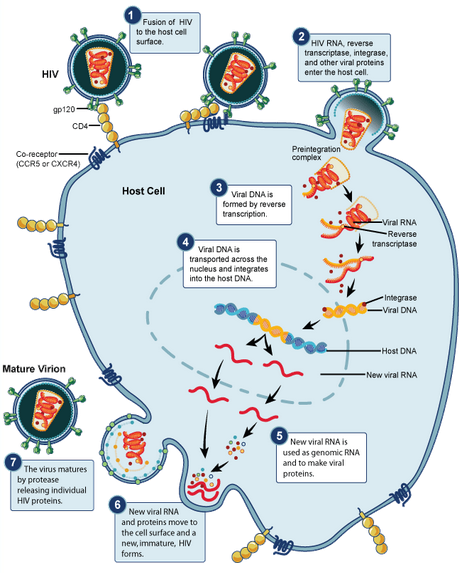
HIV begins its infection cycle when surface proteins in the phospholipid envelope interact with CD4+ T-cell CCR5 surface proteins [8, 1]. Once bound, the HIV envelope fuses with the host cell envelope, dumping the viral contents directly into the host cytoplasm [8]. Once in the cytoplasm the reverse transcriptase enzyme uses the single-stranded RNA genome as template to create a double-stranded DNA viral genome, which becomes inserted into the host genome, where it resides and safely replicates with the host cell as a provirus (figure 3) [8]. <br> | HIV begins its infection cycle when surface proteins in the phospholipid envelope interact with CD4+ T-cell CCR5 surface proteins [8, 1]. Once bound, the HIV envelope fuses with the host cell envelope, dumping the viral contents directly into the host cytoplasm [8]. Once in the cytoplasm the reverse transcriptase enzyme uses the single-stranded RNA genome as template to create a double-stranded DNA viral genome, which becomes inserted into the host genome, where it resides and safely replicates with the host cell as a provirus (figure 3) [8]. <br><br> | ||
When the host cell is no longer a suitable environment, such as during times of stress, the HIV genome replicates independently, gets transcribed into RNA, packaged into a protein capsid, and acquires its phospholipid envelope by budding out of the host cell membrane [8]. HIV has evolved the ability to bud directly to adjacent cells through cell-cell junctions, thus avoiding antibodies floating in the surrounding fluids, making it particularly effective at evading the immune system [8]. | When the host cell is no longer a suitable environment, such as during times of stress, the HIV genome replicates independently, gets transcribed into RNA, packaged into a protein capsid, and acquires its phospholipid envelope by budding out of the host cell membrane [8]. HIV has evolved the ability to bud directly to adjacent cells through cell-cell junctions, thus avoiding antibodies floating in the surrounding fluids, making it particularly effective at evading the immune system [8]. | ||
<br><b> | <br><b>History and Mutagenesis of HIV:</b><br> | ||
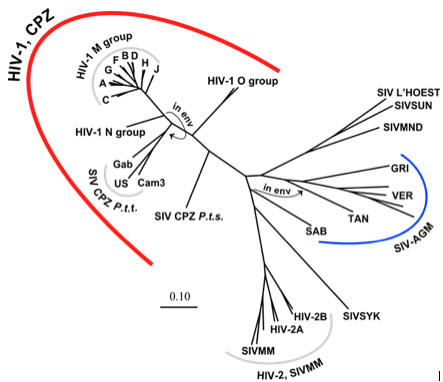
HIV is an incredibly rapidly evolving virus. It has evolved more than once, crossing the chimp-human species boundary multiple times (figure 4 [8]). While it is not usually the virus that kills the host, HIV’s invasion of T-cells weakens the immune system, allowing for opportunistic infections by other agents. The virus has the highest mutation rate known (1 in 10 base pairs) and often integrates random nucleotides when replicating, allowing the virus to continue to be unrecognizable to immune system antibodies [8]. Mutation allows for such rapid evolution of the virus that many quasi-species of HIV exist whose differences are identifiable even between human populations [8]. Because the HIV genome is double stranded, the virus has two chances of acquiring a functional genome, however with such a high mutation rate most viruses are, in fact, still defective [8]. This high rate of mutation can actually be seen as a strategy; the advantage of evading the immune system is greater for HIV than the risk of producing defective offspring. | |||
<br><b>Development of HIV-1 as a Gene Therapy Agent:</b><br> | |||
HIV-1 can be used as a gene therapy vector through the replacement of virulence genes with therapeutic transgenes. These modified viruses are allowed to infect cells, where they undergo their natural infection cycle resulting in the insertion of the therapeutic genes instead of provirus genes. <br> | |||
<br><i>Recombinant Plasmids Used for Transfection:</i><br> | |||
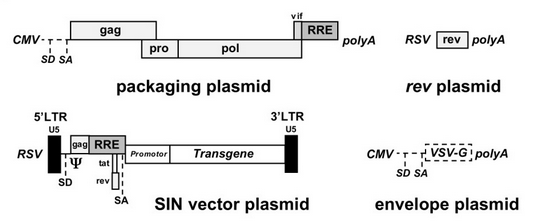
Recombinant plasmids are produced by transiently transfecting either HEK 293 or 293T cells with three plasmids containing only essential viral genes and the transgene [10]. HIV particles are allowed to infect the transfected cells, where they package and thus carry only the gene of interest. This complex process is achieved through the design and organization of at least three transfected plasmids (figure 5). <br> | |||
<br> The current lentiviral vectors used are third generation lentiviral vectors, which have been corrected to ensure safety and prevent the formation of replication competent lentiviuses (RCLs) through recombination between plasmids. Together, the plasmids contain only the three essential retroviral genes plus the transgene separated across three distinct plasmids. By deleting unnecessary proteins and allocating parts of the genome to different plasmids, it is extremely unlikely to acquire replication-competent lentiviruses [10]. <br><br> | |||
Plasmid 1 contains a promoter and viral genes gag and pol provided in trans, necessary to generate a functional virus [10]. Plasmid 2 contains the transgene acting in cis inserted into the original lentiviral genome after virulence genes vif, vpr, vpu, rev, tat, and nef have been removed from the original plasmid [10]. The transgene is located behind the packaging signal (Ψ) [10]. The use of Ψ is an important development in the production of lentiviral vectors, as any RNA molecule within 8-10 kb located behind Ψ will be packaged into the resulting lentiviruses providing the three essential gag, pol, and env genes are also provided in trans [12]. Because plasmid 2 is generated from the original lentiviral genome, the transgene and packaging signal are bordered by a modified version of the LTRs with the 3’ promoter deleted [10]. Plasmid 3 contains the third essential protein, env, encoding for the envelope glycoprotein, as well as a receptor-binding domain (SU) that can be altered to increase cellular tropism [10]. <br><br> | |||
[[Image:StructureofHIV2.png|Figure 1. Structure of the HIV virus. Source: Anderson et al 2009. http://www.nature.com/mt/journal/v17/n12/full/mt2009187a.html]] | [[Image:StructureofHIV2.png|Figure 1. Structure of the HIV virus. Source: Anderson et al 2009. http://www.nature.com/mt/journal/v17/n12/full/mt2009187a.html]] | ||
Revision as of 19:13, 23 April 2013
Introduction
What is gene therapy?
Gene therapy is a method of treating genetic disorders in which an individual’s genes are modified to correct or prevent a disease. Depending on the genetic disease, the subject’s cells can be treated by replacement of a mutant gene with a healthy gene, deactivation of a disease gene, or introduction of a gene to fight a disease. Unlike pharmaceutical drugs, gene therapy often offers a permanent cure for individuals who have inherited diseased genes, rather than treatment to manage symptoms. One mechanism of cell transformation for gene therapy involves the use of modified viral agents to introduce or remove genes from host cells.
Why use viral agents?
Viruses are naturally occurring obligate intra-cellular infectious agents that require host cell machinery in order to replicate [12]. During replication, the virus transfects the host cell with its own DNA. Researchers have taken advantage of the efficiency of viral transformation by modifying the viral genome to carry a therapeutic gene in place of a non-essential viral gene [12]. To date, viral vectors have been used to induce pluripotent stem cells, silence genes, induce transgene expression, and confer immunization [10].
What are Lentiviruses?
For some viral genera, replication involves the insertion of the viral genome into that of the host cell. Once integrated, the viral genes are replicated and passed on to daughter cells as though a part of the original host cell. One such genera is the Lentivirus genera of the Retroviridae family, who use a reverse transcriptase enzyme to produce a DNA copy of its RNA genome before inserting its DNA genome into that of the host [8]. Lentiviruses are appealing as gene therapy agents because of this permanency of transformation. Lentivuses were first proposed in 1996 as gene therapy vectors because of this ability to integrate their genome into host DNA, as well as their ability to target different cell types and infect both dividing and non-dividing cells [10]. One species of lentivirus, HIV-1, the primary cause of HIV and AIDS in humans, has been developed as a vector because of its specificity of integration at localized hotspots within the human genome, making it less likely to insert randomly and interfere with critical genes than other viral vectors, such as the murine leukemia virus [7].
Using Lentiviral Vectors to fight HIV:
One of the most encouraging advances in the use of the HIV-1 virus as a vector has actually been to fight HIV itself. Recent studies have shown that lentiviruses can be used to target key virus or host genes to confer resistance to the HIV-1 virus. This can be achieved by either targeting essential viral genes, such as replication machinery, or targeting host genes that are involved in allowing viral entry into the cell. One promising example of using HIV-1 to fight HIV involves a vector that contains three anti-HIV transgenes. Anderson et al (2009) have shown that transforming CD34+ hematopoietic progenitor cells with CCR5 short hairpin RNA gene, a TRIM5α gene, and a transactivation response element gene, each which operate at different stages of the viral life cycle, successfully block HIV infection [1].
Overview of Lentiviral Vectors
What is the HIV-1 virus?
The HIV-1 virus is the causative agent responsible for the Acquired Immunodeficiency Syndrome (AIDS) epidemic affecting roughly 30 million people worldwide [15]. The virus is transmitted between humans most commonly through unprotected sexual intercourse [4]. Once in the body, HIV infects CD4+ lymphocytes, macrophages, and dendritic cells of the immune system [4]. Being a member of the retroviral family, HIV inserts its DNA into the host genome, thus proving to be very difficult to eradicate once the infection process has begun. This leads to the destruction of CD4+ T cells and a subsequent weakening of the immune system, causing exhaustion and allowing for opportunistic infections [4].
Brief Overview of the Life Cycle of HIV-1:
The HIV-1 virus consists of a diploid single-stranded RNA genome surrounded by a protein coat and outer phospholipid envelope (figure 1, [10]). Its genome contains three essential retroviral genes as well as well as regulatory genes that modulate host interactions (figure 2). The gag gene is responsible for glycoprotein structure, the env gene transcribes envelope proteins, and the pol gene transcribes the reverse transcriptase, responsible for producing a DNA copy of the HIV RNA genome [8]. Viral genes vif, vpr, vpu, rev, nef and tat transcribe virulence proteins [10]. All proviral genes, those that become integrated into the host (gag, pol, pro, vif, vpr, vpu, tat, and env), are bordered by two long terminal repeats (LTRs) essential for gene regulation [10]. Viral promoters and enhancers are located in the LTR U3 region [10]. The packaging signal, Ψ, is located in the 5’ end of the genome, and is responsible for packaging genes downstream of itself into the viral capsid during replication [10].
HIV begins its infection cycle when surface proteins in the phospholipid envelope interact with CD4+ T-cell CCR5 surface proteins [8, 1]. Once bound, the HIV envelope fuses with the host cell envelope, dumping the viral contents directly into the host cytoplasm [8]. Once in the cytoplasm the reverse transcriptase enzyme uses the single-stranded RNA genome as template to create a double-stranded DNA viral genome, which becomes inserted into the host genome, where it resides and safely replicates with the host cell as a provirus (figure 3) [8].
When the host cell is no longer a suitable environment, such as during times of stress, the HIV genome replicates independently, gets transcribed into RNA, packaged into a protein capsid, and acquires its phospholipid envelope by budding out of the host cell membrane [8]. HIV has evolved the ability to bud directly to adjacent cells through cell-cell junctions, thus avoiding antibodies floating in the surrounding fluids, making it particularly effective at evading the immune system [8].
History and Mutagenesis of HIV:
HIV is an incredibly rapidly evolving virus. It has evolved more than once, crossing the chimp-human species boundary multiple times (figure 4 [8]). While it is not usually the virus that kills the host, HIV’s invasion of T-cells weakens the immune system, allowing for opportunistic infections by other agents. The virus has the highest mutation rate known (1 in 10 base pairs) and often integrates random nucleotides when replicating, allowing the virus to continue to be unrecognizable to immune system antibodies [8]. Mutation allows for such rapid evolution of the virus that many quasi-species of HIV exist whose differences are identifiable even between human populations [8]. Because the HIV genome is double stranded, the virus has two chances of acquiring a functional genome, however with such a high mutation rate most viruses are, in fact, still defective [8]. This high rate of mutation can actually be seen as a strategy; the advantage of evading the immune system is greater for HIV than the risk of producing defective offspring.
Development of HIV-1 as a Gene Therapy Agent:
HIV-1 can be used as a gene therapy vector through the replacement of virulence genes with therapeutic transgenes. These modified viruses are allowed to infect cells, where they undergo their natural infection cycle resulting in the insertion of the therapeutic genes instead of provirus genes.
Recombinant Plasmids Used for Transfection:
Recombinant plasmids are produced by transiently transfecting either HEK 293 or 293T cells with three plasmids containing only essential viral genes and the transgene [10]. HIV particles are allowed to infect the transfected cells, where they package and thus carry only the gene of interest. This complex process is achieved through the design and organization of at least three transfected plasmids (figure 5).
The current lentiviral vectors used are third generation lentiviral vectors, which have been corrected to ensure safety and prevent the formation of replication competent lentiviuses (RCLs) through recombination between plasmids. Together, the plasmids contain only the three essential retroviral genes plus the transgene separated across three distinct plasmids. By deleting unnecessary proteins and allocating parts of the genome to different plasmids, it is extremely unlikely to acquire replication-competent lentiviruses [10].
Plasmid 1 contains a promoter and viral genes gag and pol provided in trans, necessary to generate a functional virus [10]. Plasmid 2 contains the transgene acting in cis inserted into the original lentiviral genome after virulence genes vif, vpr, vpu, rev, tat, and nef have been removed from the original plasmid [10]. The transgene is located behind the packaging signal (Ψ) [10]. The use of Ψ is an important development in the production of lentiviral vectors, as any RNA molecule within 8-10 kb located behind Ψ will be packaged into the resulting lentiviruses providing the three essential gag, pol, and env genes are also provided in trans [12]. Because plasmid 2 is generated from the original lentiviral genome, the transgene and packaging signal are bordered by a modified version of the LTRs with the 3’ promoter deleted [10]. Plasmid 3 contains the third essential protein, env, encoding for the envelope glycoprotein, as well as a receptor-binding domain (SU) that can be altered to increase cellular tropism [10].

At right is a sample image insertion. It works for any image uploaded anywhere to MicrobeWiki. The insertion code consists of:
Double brackets: [[
Filename: PHIL_1181_lores.jpg
Thumbnail status: |thumb|
Pixel size: |300px|
Placement on page: |right|
Legend/credit: Electron micrograph of the Ebola Zaire virus. This was the first photo ever taken of the virus, on 10/13/1976. By Dr. F.A. Murphy, now at U.C. Davis, then at the CDC.
Closed double brackets: ]]
Other examples:
Bold
Italic
Subscript: H2O
Superscript: Fe3+
Overview of Lentiviral Vectors
Section 2
Include some current research in each topic, with at least one figure showing data.
Section 3
Include some current research in each topic, with at least one figure showing data.
Conclusion
Overall paper length should be 3,000 words, with at least 3 figures.
References
Edited by student of Joan Slonczewski for BIOL 238 Microbiology, 2009, Kenyon College.
