Dehalobacter: Difference between revisions
| Line 30: | Line 30: | ||
The whole genome of Dehalobacter restrictus PER- K23 was recently sequenced revealing critical insights into phylogeny and metabolic capabilities of the genus. The 2,943 kbp long genome contains 2,826 protein coding and 82 RNA genes, including 5 16S rRNA genes. Interestingly, the genome contains 25 predicted rdase genes, the majority of which appear to be full length. The rdase genes are mainly located in two clusters, suggesting a much larger potential for organohalide respiration than previously anticipated (11). | The whole genome of Dehalobacter restrictus PER- K23 was recently sequenced revealing critical insights into phylogeny and metabolic capabilities of the genus. The 2,943 kbp long genome contains 2,826 protein coding and 82 RNA genes, including 5 16S rRNA genes. Interestingly, the genome contains 25 predicted rdase genes, the majority of which appear to be full length. The rdase genes are mainly located in two clusters, suggesting a much larger potential for organohalide respiration than previously anticipated (11). | ||
==Cell Structure | ==Cell Structure== | ||
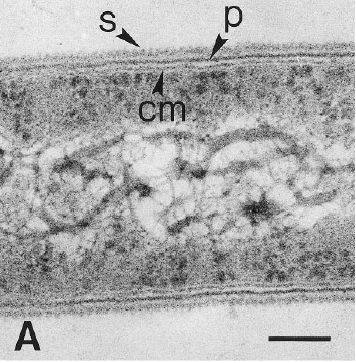
[[Image:close_up.GIF|frame|left|Electron micrograph of a thin-section of strain PER-K23, showing the cytoplasmic membrane (cm), the peptidoglycan layer (p), and the surface layer (s). From [http://lbewww.epfl.ch/LBE/PDF/Publi/CH5.pdf Holliger ''et al''.]]] | [[Image:close_up.GIF|frame|left|Electron micrograph of a thin-section of strain PER-K23, showing the cytoplasmic membrane (cm), the peptidoglycan layer (p), and the surface layer (s). From [http://lbewww.epfl.ch/LBE/PDF/Publi/CH5.pdf Holliger ''et al''.]]] Dehalobacter restrictus cells are rod shaped, 2-3 µm in length and 0.3-0.5 µm in diameter with tapering ends. There is a single lateral flagellum at one end indicating that the cells are motile. The cells do not form spores under substrate and temperature induced stress. Cell wall stains gram negative and is composed of peptidoglycan type A3ϒ enveloped by a layer of proteinaceous surface layer (S – layer) (5) | ||
[[Image:dehalogenation.GIF|frame|center| Reductive dechlorination of chloroethenes, starting with tetrachloroethene (PCE), to trichloroethene (TCE), to dichloroethene (represented by ''cis''-DCE), to vinyl chloride (VC), to ethene (ETH). | [[Image:dehalogenation.GIF|frame|center| Reductive dechlorination of chloroethenes, starting with tetrachloroethene (PCE), to trichloroethene (TCE), to dichloroethene (represented by ''cis''-DCE), to vinyl chloride (VC), to ethene (ETH). | ||
Revision as of 18:52, 18 April 2014
A Microbial Biorealm page on the genus Dehalobacter
Classification
Higher order taxa:
Bacteria; Firmicutes; Clostridia; Clostridiales; Peptococcaceae; Dehalobacter
Species:
Dehalobacter restrictus PER K23, Dehalobacter sp. E1, Dehalobacter sp. TCA1, Dehalobacter sp. MS Dehalobacter sp. WL
|
NCBI: Taxonomy Genome |
Description and Significance
Dehalobacter restrictus is a rod shaped gram-negative obligate anaerobe found in deep soil sediments and groundwater aquifers and is phylogenetically related to several halorespiring genera like Desulfitobacterium and Desulfosporlosinus. It is a typical representative of Dehalobacter spp., which are capable of reductive dechlorination of chlorinated ethenes and chlorinated ethanes (1, 2) and fermentative dehalogenation of Dichloromethane (DCM) (3). These metabolic pathways make them relevant to bioremediation of ground water aquifers contaminated with chlorinated solvents (4). To date, several strains of Dehalobacter have been isolated (1, 3, 5-7)."
Genome Structure
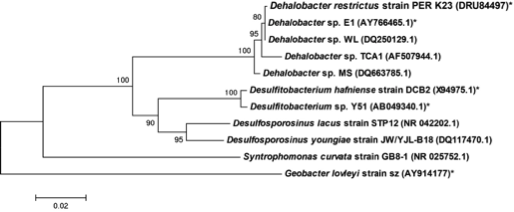
The whole genome of Dehalobacter restrictus PER- K23 was recently sequenced revealing critical insights into phylogeny and metabolic capabilities of the genus. The 2,943 kbp long genome contains 2,826 protein coding and 82 RNA genes, including 5 16S rRNA genes. Interestingly, the genome contains 25 predicted rdase genes, the majority of which appear to be full length. The rdase genes are mainly located in two clusters, suggesting a much larger potential for organohalide respiration than previously anticipated (11).
Cell Structure
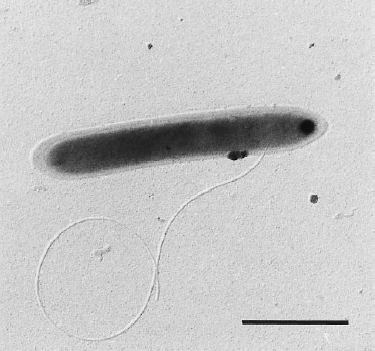
Dehalobacter restrictus cells are rod shaped, 2-3 µm in length and 0.3-0.5 µm in diameter with tapering ends. There is a single lateral flagellum at one end indicating that the cells are motile. The cells do not form spores under substrate and temperature induced stress. Cell wall stains gram negative and is composed of peptidoglycan type A3ϒ enveloped by a layer of proteinaceous surface layer (S – layer) (5)
[[Image:dehalogenation.GIF|frame|center| Reductive dechlorination of chloroethenes, starting with tetrachloroethene (PCE), to trichloroethene (TCE), to dichloroethene (represented by cis-DCE), to vinyl chloride (VC), to ethene (ETH).
Ecology
Tetrachloroethene (PCE), also known as perchloroethene and tetrachloroethylene, is a volatile chlorinated hydrocarbon, widely used as a lipophilic solvent, and is one of the most abundant halogenated pollutants. It is widely used for dry cleaning of textiles and degreasing of machine and metal parts. PCE can dissolve fats, greases, waxes and oils off of both man-made and natural fibers, as well as metal parts and other fabricated materials. It is very prevalent in the environment due to poor handling and its resistance to aerobic transformation. It has been released into the environment for decades and has become one of the most common contaminates of soils and groundwater.
Although the government has banned land disposal, PCE is still released into the environment in alarming quantities. In 1993 it is estimated that 11.2 million pounds of PCE was released into the air, and 10,152 pounds was released into the water. Although these numbers are down from previous years, concentrations in the environment are still too high to be considered safe.
PCE is considered a carcinogen, although there is little evidence to truly substantiate this claim, all lightweight chlorinated hydrocarbons are treated as potential carcinogens (although some, such as vinyl chloride, are better documented than others). The carcinogenicity of PCE comes from the interaction of the chemical metabolites with DNA. This occurs when the body attempts to create a more water-soluble compound that can be processed by the kidneys in order to get rid of it. This process usually takes two steps, the formation of an epoxide intermediate, which is then metabolized into a diol, which can be excreted by the body. Epoxides react very aggressively with DNA, and even though they are only around for a very short time, they can create mutations, and when these mutations hit oncogenes, cancer can be induced.
As mentioned above, Dehalobacter restrictus performs anaerobic respiration using PCE. In a process called dehalorespiration D. restrictus respires by oxidation of dihydrogen (using it as an electron donor) and reductive dechlorination of PCE (uses it as an electron acceptor). PCE is produced naturally in the environment by several species of temperate and subtropical marine algae. For the purpose of disposing of PCE, Dehalobacter restrictus is very promising.
Isolation and Cultivation
D. restrictus is isolated from a laboratory system that was being fed with tetrachloroethene.