Teixobactin
The development of new antibiotics has been a difficult task due to the rapid evolution of resistant bacteria. Teixobactin is a newly discovered antibiotic that is effective against gram positive bacteria including antibiotic-resistant strains, such as methicilin-resistant Staphylococcus aureus (MRSA) and vancomycin-resistant enterococci (VRE), without evidence of resistance development.1 The antibiotic, isolated from the soil using a device called the iChip, works by inhibiting cell wall synthesis by binding to two lipid cell wall precursors.1
Introduction
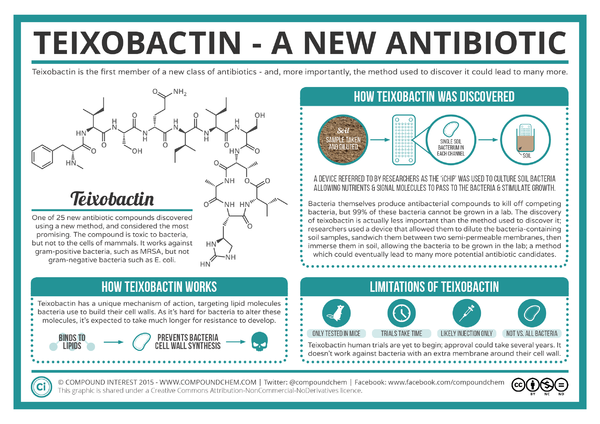
Microorganisms have been our primary source of new antibiotics since the drugs' introduction in the 1940s.1 However, only about 1% of all bacterial species are readily culturable in laboratory conditions, and the ability of microorganisms to rapidly develop drug resistance makes the the search for new antibiotics a difficult process.1 NovoBiotic Pharmaceuticals in collaboration with the University of Bonn and Northeastern University reported in Nature the discovery of a new antibiotic, teixobactin, with properties that minimize the development of bacterial resistance.3 Ling et al. found teixobactin to be effective against gram positive bacteria, including drug-resistant strains such as MRSA and VRE. This antibiotic, isolated using a device called the iChip, works through inhibiting cell wall synthesis by binding to
Overall paper length should be 3,000 words, with at least 3 figures with data.
Identification
Teixobactin was isolated from a new species, Eleftheria terrae, from soil using a high-throughput device called the iChip.1 The iChip is used to isolate previously unculturable bacteria by growing microbes in situ where their normal environmental factors are used to cultivate the bacteria in hundreds of miniature diffusible chambers on the iChip.2 Compared to standard Petri dishes, the colony count for soil bacteria using the iChip was 5 times higher when cultivating from a single colony's worth of bacteria.2 Extracts from thousands of isolates were tested for antibiotic activity, and the discovery of teixobactin's properties lead to the identification and classification of E. terrae using 16S rRNA gene sequencing.1 Relatives of E. terrae were not previously known to produce antibiotics.1 Though the use of the iChip's cultivating abilities, other antibiotics like teixobactin could be discovered.
Mechanism of Action, In Vivo Efficacy, and Resistance of Teixobactin
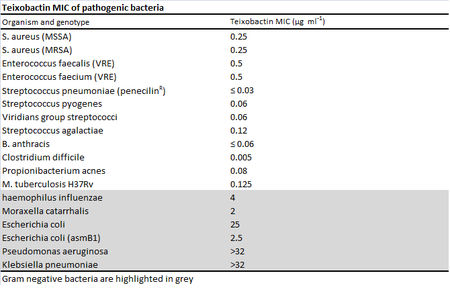
Minimum inhibitory concentrations (MIC) were used to measure the effectiveness of teixobactin against pathogenic bacteria.1 While teixobactin was not effective against gram-negative bacteria, less than 0.6 μg/mL of the drug was necessary to visibly inhibit the growth of many gram positive bacteria tested including pathogenic MRSA, VRE, Bacillus anthracis, and Clostridium difficile.1 No toxicity toward mammalian cells was found as expected from the mechanism of action . The antibiotic works through inhibiting cell wall synthesis by binding to two cell wall precursors, lipid II and lipid III. 1 Though researchers looked for possible teixobactin-resistant mutants using S. aureau and M. tuberculosis through plating the bacteria in media with four times their respective MICs, no resistant mutants were found.1 Studies done in the past on vancomycin, another antibiotic that binds to lipid cell wall precursors, suggest that teixobactin's lack of development of resistance may be due to the fact that the lipids are made from organic precursors rather than synthesized de novo from DNA like proteins, making development of resistance through mutation alone more difficult.4 In addition, the binding target of the drug is highly conserved among bacteria.1 Resistance to vancomycin was identified almost 40 years after the drug's discovery when it is believed that the self-resistance vector from vanomycin-producing bacteria was captured by pathogenic bacteria through horizontal gene transfer.4
Limitations of Teixobactin
Although resistance to teixobactin was difficult to manufacture in lab, resistance could eventually emerge in the same way vancomycin resistance emerged, through horizontal gene transfer.1 However, as E. terrae is gram negative and does not need carry genes for self-resistance like vancomycin-producing bacteria, the genes for resistance would likely come from other soil bacteria.1
Further Reading
[Sample link] Ebola Hemorrhagic Fever—Centers for Disease Control and Prevention, Special Pathogens Branch
References
Edited by (your name here), a student of Nora Sullivan in BIOL168L (Microbiology) in The Keck Science Department of the Claremont Colleges Spring 2014.
