Vibrio cholerae pathogenesis
Vibrio cholerae is the causative agent of the diarrheal disease cholera. A disease affecting 2.8 million people per year and resulting in the deaths of 91,000, cholera is most common in areas with high population density and low sanitation quality.[1] [2] Not all V. cholerae are pathogenic: only two strains of serogroups O1 and O139 cause cholera.[1] Furthermore, it is considered a facultative human pathogen, as it primarily inhabits surface waters, as opposed to the small intestine.[3]
When entering and colonizing the human host, V. cholerae must endure changing environmental factors such as temperature, acidity, osmolarity, intestinal growth inhibitory substances, and immune system factors.[1] Toxin-coregulated pilus (TCP) is then necessary for colonization of the small intestine, while cholera toxin (CT) is necessary for the watery diarrhea response.[4] Understanding pathogenesis of V. cholerae requires an understanding of colonization and these virulence factors.
Colonization of the Human Small Intestine
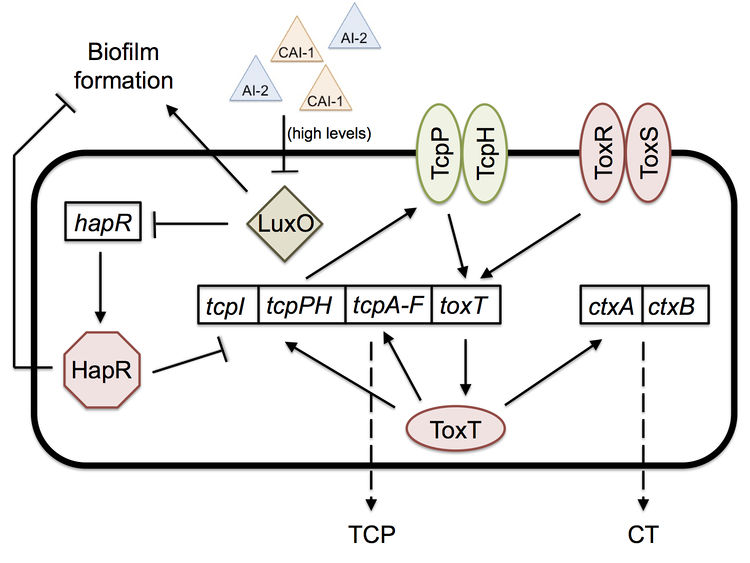
Biofilm Formation
V. cholerae’s journey through the acidic stomach is aided by its assembly into a biofilm, a structure consisting of bacteria in a matrix of sugars and proteins.[1] [3] 100% of V. cholerae cells in biofilms have been shown to withstand acid shock characteristic of the human stomach. Exposure to an environment with a pH of 4.5 for 15 and 30 minutes improves cell survival 50 and 1000-fold, respectively, compared to non-biofilm-associated cells.[5]
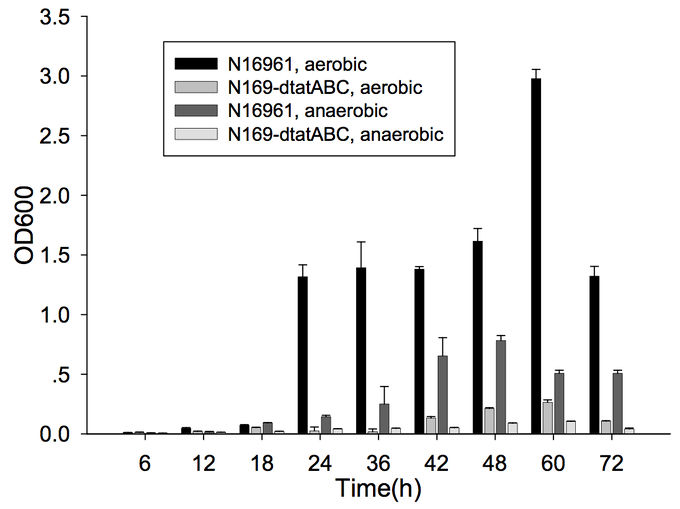
Furthermore, cells capable of forming biofilms have greater intestinal colonization capacity in the suckling mouse model. The rugose variant of V. cholerae, which has superior biofilm-forming abilities compared to the smooth variant, is also more successful at colonizing the intestine.[6] In addition, the twin-arginine translocation (Tat) system has also been shown to be important for biofilm formation and for colonization. The Tat system is an export system that recognizes the twin arginine signal peptide on proteins that are intended to be translocated across the cell membrane (these are often enzymes involved in the electron transport chain). This system has already been associated with virulence of Pseudomonas aeruginosa, Escherichia coli, Yersinia pseudotuberuclosis, and other pathogenic bacteria. The Tat system has recently been shown to be functional in V. cholerae; deletion of tatABC causes a decrease in biofilm formation (Figure 1) as well as in colonization of the suckling mouse.[7] Finally, the absence of vibrio polysaccharides (encoded by the vps operon), the major exopolysaccharide component of biofilms, or the absence of RbmA, one of the matrix proteins, leads to defective colonization.[6]
However, quorum sensing studies in V. cholerae have shown that the biofilm is not necessary during colonization, but instead during the bacteria’s travel through the acidic stomach, during which they need a protective structure. Once they reach the intestine, they will then utilize virulence factors, not the biofilm, to colonize and infect the host.
Quorum Sensing
Once in the intestine, the biofilm is no longer of help to V. cholerae, and a new mechanism is necessary for colonization. Quorum sensing, a bacterial mechanism of communication with downstream effects on gene regulation based on population density, is what allows the bacteria to colonize the intestine by expressing virulence factors, such as cholera toxin (CT) and the toxin-coregulated pilus (TCP).[8]
Two important quorum-sensing related proteins of note are LuxO and HapR, which work together in a system similar to that of Vibrio harveyi, the most common bacterium used in quorum sensing studies.[9] LuxO is a regulator that takes the information provided by levels of autoinducer 2 (AI-2) and cholera autoinducer 1 (CAI-1) – detected by LuxPQ and CqsS membrane receptors, respectively – and represses the master transcription factor HapR.[10] [11] In V. harveyi, this activation of LuxO and repression of HapR leads to a decrease in luminescence (as HapR in the bacterium transcriptionally activates the luciferase operon). V. cholerae does not have a bioluminescence system and HapR transcriptionally represses virulence genes.[11] When LuxO is active, it represses expression of hapR, thus allowing the transcription of tcpP, a gene necessary for the biosynthesis (though not itself a building block) of the TCP (Figure 2). CT (responsible for the diarrhea characteristic of cholera) expression is dependent upon the presence of TCP, so active LuxO and inactive HapR result in an increased amount of both TCP and CT.[11] LuxO and HapR’s effect on virulence has been demonstrated in the CD-1 suckling mouse model: deletion of luxO results in no cell recovery from the intestine of the biopsied mouse (no colonization occurred), while deletion of hapR does not affect virulence and colonization at all.[11] Furthermore, toxigenic strains of V. cholerae have hapR mutations, which may serve to improve their colonization of humans by increasing TCP and CT levels.
Quorum sensing and biofilm formation have been shown to have a reciprocal relationship in V. cholerae, unlike in most other biofilm-forming species.[5] The current working model of colonization is that the biofilm is necessary to protect V. cholerae as it goes through the acidic stomach environment, since it has been shown to increase tolerance to acid shock.[5] However, once the bacterium is in the small intestine, in which it no longer needs the biofilm’s protection, the high density of cells causes high levels of quorum sensing autoinducers. High levels of the autoinducer CAI-1 represses LuxO and activates HapR, which then represses transcription of the vps operon, thus preventing further biofilm formation and improving the bacterium’s colonization ability; hapR mutants have shown to have thicker biofilms with 10-fold lower colonization in suckling mice.[5] The cells disperse from the biofilm and as a result have lower levels of autoinducers. Low levels allow the activation of LuxO, the repression of HapR, and thus the expression of tcpP and the ctx operon.[5] [11] Production of TCP and CT allow for infection to occur. Once enough V. cholerae accumulate throughout the infection, high levels of autoinducers will repress TCP and CT production, thus activating protease production, which leads to the detachment of bacteria from the intestinal mucosa and their exit from the host.[5]
Toxin-Coregulating Pilus
Bacterial pili are typically involved in surface motility, microcolony formation, biofilm formation, host cell adhesion, cell signaling, DNA uptake, and phage attachment. They are usually very strong, withstanding forces of 100 pN or more. Toxin-coregulating pilus (TCP) is the pilin for V. cholerae, considered very important for its virulence.[12]
Structure
TCP is classified as a type IVb pilin (i.e. it is present on bacteria that colonize the human intestines), which are typically 1-4 μm long, 50-80 Å thick, and flexible. TCP is composed of many copies of its subunit, TcpA, a 199 amino acid polypeptide, the largest type IV pilin subunit.[13]
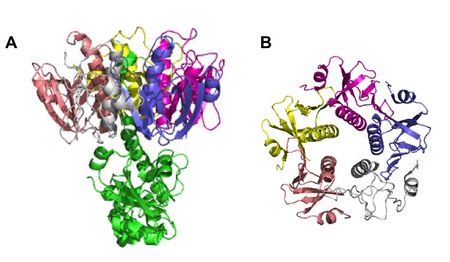
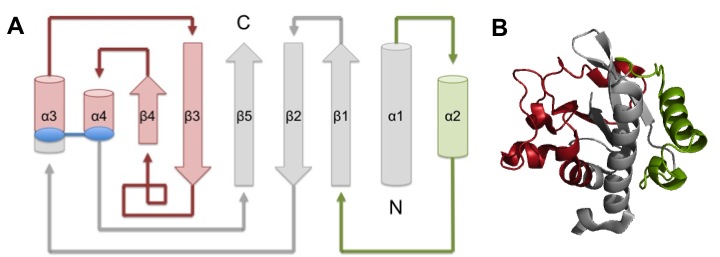
TcpA is composed most notably of a globular head domain and a D-region (Figure 3). The globular head domain consists of the N-terminal α-helix, a second shorter α-helix, and an antiparallel β-sheet; together, they form a tight hydrophobic core. The head has two sides: one is the loop connecting the N-terminal α-helix to the β-sheet, called the αβ-loop, and the other is the D-region. The D-region is composed of 2 α-helices and 2 antiparallel beta sheets. It is responsible for interacting with the other TcpA subunits to form large TCP and stabilize the whole structure with disulfide bonds.[13]
Assembly of the TCP structure is organized in 3 identical protein strands which twist around each other to form a helical twist filament. The filament has axial separation between the strands of 45 Å and each turn of the filament has 6 TcpA subunits per strand. There is a polar interface between TcpA along the fiber axis, which binds the 3 strands together. There is also a hydrophobic interface between subunits radially.[13]
Role in Virulence
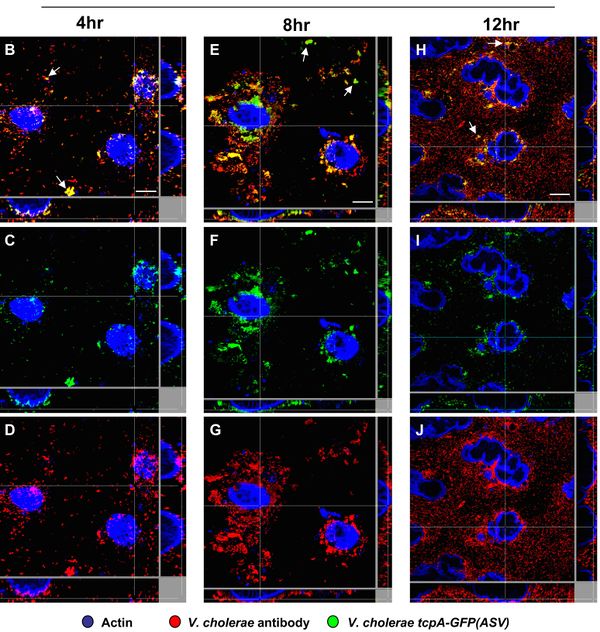
TCP, although not involved in causing the symptoms of cholera, has been shown to be absolutely necessary for colonization and subsequent infection by V. cholerae. tcpA is required for colonization of both CD-1 mice and humans. Although tcpA deletion mutant cells can grow in vitro without TCP, they are unable to colonize mice, as seen by analysis of intestinal homogenates.[14] In humans, tcpA mutant cells cause no clinical response and are not found in stool samples (while wild type cells are).[15] Although not yet proven, it is hypothesized this is due to TCP’s role in adherence to the mucosal surface, which TCP may mediate by using bacterial adhesions, which would bind epithelial cell surface receptors and create microcolonies on intestinal epithelia. Research has shown that TcpA increases the surface hydrophobicity of the cells, which may cause autoagglutination in liquid culture and microcolonies in the intestine. [16] This concentration of cells on the mucosal surface is what allows the next step of virulence, delivery of CT, to occur (once it reaches 107-8 cells per gram of mucosal surface).[17] Without tcpA, CT levels are decreased by half in the mouse intestine.[14] This decrease is suspected to be caused by in vivo effects of the mutation, rather than downstream transcriptional changes, as tcpA mutants do not affect CT levels in vitro as well. It is hypothesized that TCP increasing microcolony formation on the intestinal epithelia allows intestinal signals to reach the cell and activate virulence gene (such as ctx) expression.[18]
tcpA has also been shown to have temporal and spatial regulation in mouse and rabbit models. It is expressed in the small intestine with two distinct temporal and spatial patterns. It is first expressed when V. cholerae is in the lumen of the upper GI tract, but at very low levels; this is suspected to cause some TCP assembly. Then, it is expressed in more abundance in the small intestine.[18] Within the small intestine, there is an increasing gradient of tcpA expression from the mucus gel toward the surface of the epithelial tissue (Figure 4).[19]
Cholera Toxin
Although hypothesized to exist back in 1884, V. cholerae’s toxin, cholera toxin (CT), was not discovered until 1959, when it was found in rabbit culture filtrates.[3] In the next 15 years, it was purified and sequenced and found to be composed of six subunits in total: five binding subunits (encoded by ctxB) and one active subunit (ctxA) (Figure 5).[20] The B subunits are 103 amino acids each, while the A subunit is 240 amino acids.[21] As a whole, CT is 84,000 kDa.[20]
CT’s binding of the GM1 ganglioside was originally hypothesized when there appeared to be a relationship between the ganglioside concentration, the number of binding sites for CT, and the resulting intestinal sensitivity to CT.[22] This hypothesis was confirmed when isolated CT was shown to be fixed and precipitated by the ganglioside and not any of its similar glycolipids.[23] Once the B subunits bind 5 GM1 molecules (located in the plasma membrane of the mucosal cells), the the receptor-toxin complex gets endocytosed and transported to the endoplasmic reticulum. Not all 5 subunits must bind the GM1 gangliosides for the complex to form and endocytosis to occur: while there is a decrease in toxicity with a decrease in binding, only one GM1 needs to be bound for CT to enter the host cell (Figure 6). Once in the ER, the disulfide bond in the A subunit then gets reduced, causing the A subunit to dissociate from the complex and enter the cytoplasm, where it binds an ADP ribosylation factor.[21] This process causes the activation of adenylate cyclase, which produces cyclic adenosine monophosphate (cAMP) from ATP.[24]
The increase in cAMP changes the transmural ion flux, causing the secretion of Cl- by the intestinal gland in the lining of the small intestine as well as the inhibition of neutral NaCl absorption by intestinal villi.[25] Because of this outpouring of Cl-, and the intestine’s inability to reabsorb it, the intestine’s volume reaches abnormal levels and results in the watery diarrhea characteristic of cholera. The excessive diarrhea causes the infected human to lose electrolyte-rich fluids and thus become dehydrated.[25] [26]
Future Directions in Vaccine Development
Cholera has been a burden to human health for centuries and has spread globally in the past 200 years.[27] For the past 15 years, the World Health Organization has called for the development of oral cholera vaccines.[28] Options include whole-cell killed vaccines, which cause an immune response but no disease, and live-attenuated vaccines, which are weakened bacteria that cannot cause full-fledged disease, but can cause an immune response. While previously, researchers focused on creating vaccines using inactivated CT, it is now understood through animal model and human studies that anti-CT immunity that developed from treatment by the vaccine is not nearly as effective at protecting the patient from cholera. Finally, one could use outer membrane vesicles released by V. cholerae or synthetic liposomes to deliver cholera antigens.[29] The next few decades should show further development and clinical trials of these various vaccines.
References
[1] Reidl, J. and K.E. Klose, Vibrio cholerae and cholera: out of the water and into the host. Fems Microbiology Reviews, 2002. 26: p. 125-139.
[2] Ali, M., et al., The global burden of cholera. Bulletin of the World Health Organization, 2012. 90: p. 209-218.
[3] Sack, D.A., et al., Cholera. Lancet, 2004. 363: p. 223-233.
[4] Yu, R.R., and V.J. DiRita, Analysis of an autoregulatory loop controlling ToxT, cholera toxin, and toxin-coregulated pilus production in Vibrio cholerae. Journal of Bacteriology, 1999. 181: p. 2584-2592.
[5] Zhu, J. and J.J. Mekalanos, Quorum sensing-dependent biofilms enhance colonization in Vibrio cholerae. Developmental Cell, 2003. 5: p. 647-656.
[6] Fong, J.C.N., et al., Role of Vibrio polysaccharide (vps) genes in VPS production, biofilm formation and Vibrio cholerae pathogenesis. Microbiology, 2010. 156: p. 2757-2769.
[7] Zhang, L., et al., Pleiotropic effects of the twin-arginine translocation system on biofilm formation, colonization, and virulence in Vibrio cholerae. Biomed Central Microbiology, 2009. 9: p. 1-13.
[8] Camilli, A. and B.L. Bassler, Bacterial small-molecule signaling pathways. Science, 2006. 311: p. 1113-1116.
[9] Miller, M.B., et al., Parallel quorum sensing systems converge to regulate virulence in Vibrio cholerae. Cell, 2002. 110: p. 303-314.
[10] Higgins, D.A., et al., The major Vibrio cholerae autoinducer and its role in virulence factor production. Nature, 2007. 450: p. 883-886.
[11] Zhu, J., et al., Quorum-sensing regulators control virulence gene expression in Vibrio cholerae. Proceedings of the National Academy of Sciences USA, 2002. 99: p. 3129-3134.
[12] Craig, L., et al., Type IV pilus structure and bacterial pathogenicity. Nature Reviews Microbiology, 2004. 2: p. 363-378.
[13] Craig, L., et al., Type IV pilin structure and assembly: X-ray and EM analyses of Vibrio cholerae toxin-coregulated plius and Pseudomonas aeruginosa PAK pilin. Molecular Cell, 2003. 11: p. 1139-1150.
[14] Thelin, K.H. and R.K. Taylor, Toxin-coregulated pilus, but not mannose-sensitive hemagglutinin, is required for colonization by Vibrio cholerae O1 El Tor biotype and O139 strains. Infection and Immunity, 1996. 64: p. 2853-2856.
[15] Herrington, D.A., et al., Toxin, toxin-coregulated pili, and the toxR regulon are essential for Vibrio cholerae pathogenesis in humans. Journal of Experimental Medicine, 1988. 168: p. 1487-1492.
[16] Taylor, R.K., et al., Use of phoA gene fusions to identify a pilus colonization factor coordinately regulated with cholera toxin. Proceedings of the National Academy of Sciences USA, 1987. 84: p. 2833-2837.
[17] Sprinz, H., et al., Biopsy of small bowel of Thai people with special reference to recovery from Asiatic cholera and to an intestinal malabsorption syndrome. American Journal of Clinical Pathology, 1962. 38: p. 43-51.
[18] Lee, S.H., et al., Regulation and temporal expression patterns of Vibrio cholerae virulence genes during infection. Cell, 1999. 99: p. 625-634.
[19] Nielsen, A.T., et al., A bistable switch and anatomical site control Vibrio cholerae virulence gene expression in the intestine. Public Library of Science Pathogens, 2010. 9: e1001102.
[20] Lonnroth, I. and J. Holmgren, Subunit structure of cholera toxin. Journal of General Microbiology, 1973. 76: p. 417-427.
[21] Jobling, M.G., et al., A single native ganglioside GM1-binding site is sufficient for cholera toxin to bind to cells and complete the intoxication pathway. mBio, 2012. 3: e00401-12.
[22] Holmgren, J., et al., Interaction of cholera toxin and membrane GM1 ganglioside of small intestine. Proceedings of the National Academy of Sciences USA, 1975. 72: p. 2520-2524.
[23] Holmgren, J., et al., Tissue receptor for cholera exotoxin: postulated structure from studies with GM1 ganglioside and related glycolipids. Infection and Immunity, 1973. 8: p. 208-214.
[24] Van Heyningen, W.E., et al., Deactivation of cholera toxin by ganglioside. Journal of Infectious Diseases, 1971. 124: p. 415-418.
[25] Field, M., et al., Effect of cholera enterotoxin on ion transport across isolated ileal mucosa. Journal of Clinical Investigation, 1972. 51: p. 796-804.
[26] Speelman, P., et al., Colonic dysfunction during cholera infection. Gastroenterology, 1986. 50: p. 1164-1170.
[27] Sack, D.A., et al., Getting serious about cholera. New England journal of Medicine, 2006. 355: p. 649-651.
[28] Cholera vaccines. Weekly Epidemiology Report, 2001. 76: 117-124.
[29] Vibrio cholerae: lessons for mucosal vaccine design. Expert Review Vaccines, 2011. 10: p. 79-94.
Edited by Tina Solvik, a student of Suzanne Kern in BIOL168L (Microbiology) in The Keck Science Department of the Claremont Colleges Spring 2015.