Shigella boydii
A Microbial Biorealm page on the genus Shigella boydii
Classification
Higher order taxa
Bacteria (domain); Proteobacteria (phylum); Gamma Proteobacteria (class); Enterobacteriales (order); Enterobacteriaceae (family); Shigella (genus)
Species
Shigella boydii
|
NCBI: Taxonomy |
Description and significance
In the 1950's Shigella was accepted as a genus and subgrouped into four species: S. dysenteriae, S. Fexneri, S. boydii, and S. sonnei (also referred to as subgroups A-D).[3]
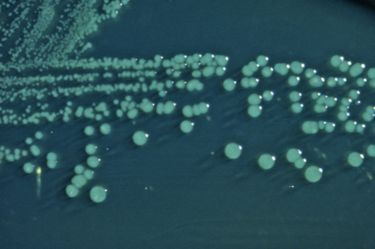
Appearance: Shigella boydii is a bacillary (rod-shaped) gram negative bacterium that does not form spores and is usually nonmotile.[2][5]
Habitat: S. boydii inhabits the inestine and rectum of humans and other primates. It can survive temporarily in feces and/or food/water contaminated with fecal matter. Locationally it is found mainly on the sub-continent of India.[3]
Significance: Human pathogen that causes bacillary dysentery.
Serotypes: There are 20 S. boydii serotypes.[5]
Genetics: Shigella bacteria are thought to be derived from different strains of Escherichia coli. S. boydii is the most genetically diverse and some serotypes seem to be more closely related to other species. S. boydii type 13, for example, shares sequence similarities with Vibrio cholerae for the genes encoding the O antigen, the polysaccharide part of the lipopolysaccharide (LPS), and therefore these may be more closely related. LPS is an endotoxin and part of the Gram-negative outer membrane.[1]
Genome structure
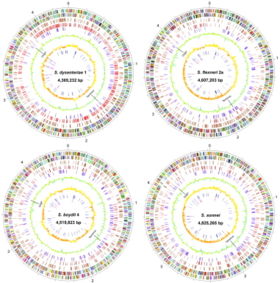
Size of genome: Shigella boydii (Sb227) serotype 4- Circular DNA chromosome with 4,519,823 nt (446 genes); Plasmid (pSB4_227)- 126,697 nt (149 genes). Both were completed 2005/11/18. [11][12]
Pathogenic Contents: The one plasmid, pSb4_227, has post-segregation killing systems mvpA/mvpT, found in all species' plasmids and ccdA/ccdB not found in some.
It is believed that most Shigella have gained and lossed Stx genes at one time or another, the toxin genes found in S. dysenteriae. One evidence of this is Sb227 which has a T2SS pseudogene. T2SS, found in S. dysenteriae, enables export of toxins to host that would otherwise build up inside of cell and cause lysis and therefore T2SS greatly contributes to pathogenicity. This inactivation of T2SS in S. boydii has most likely been naturally selected to better adapt to a human host as having greater pathogenicity offers little benefit for longterm survival.
Insertion (IS) element ISSbo6 is restricted in SHI-1, SHI-2 and ipaH pathogenicity islands. This suggestes that these islands were aquired before the IS-element.[3]
Iron Up-take: Iron is taken up from the environment by ABC transporters and by the iron transport compound aerobactin.[3]
Interesting Features: Shigella has a very large number of IS-elements which not only lead to rearrangement of DNA but also deletion. For example, the cell entry region, found in all Shigella virulence plasmids, and the icsA gene were deleted from pSB4_227. Also about 30kb that lie between icsA and ccdA in pCP301 (plasmid of S. flexneri 2a, strain 301) are missing.
All Shigella species including most of S. boydii serotypes share about 3 Mb worth of genetic material with E. coli. This supports the idea that Shigella species diverged from the same origins as E. coli.[3]
Cell Structure and Metabolism
Cell Structure: Shigella cells contain a few layers of peptidogylcan, a periplasm and a secondary outer membrane covered with lipopolysaccharides, an endotoxin that also provides structural support. These are all general characteristics of a Gram-negative bacteria. Most research would agree that Shigella are nonmotile but some evidence suggests that they do in fact have flagella, although motility is not necessary for infection of colon. The flegella tends to be on one pole of the cell and about 10 microns in length and 12- 14nm in diameter. The genes coding for the flagella in S. dysenteriae, S. flexneri, S. boydii and S. sonnei were different; attributing to the genetic diversity among the species.[8]
Metabolism: S. boydii typically does not have oxidase enzymes but rather catalase enzymes (catalyzes the reduction of H2O2->H20). The organic molecule Indole is variable. Methyl red testing is positive, meaning that the bacteria uses a mixed acid fermentation pathway. Voges–Proskauer and Simmons’ citrate reactions are negative, meaning that this organism does not utilize the butylene glycol pathway or produce acetoin. Lysine decarboxylase, arginine dihydrolase and ornithine decarboxylase are not present. S. boydii does not produce H2S, does not hydrolyze urea and does not grow in KCN broth. Carbohydrates are usually fermented and these include glucose (in the absence of gas production), D-mannitol, arabinose, trehalose and mannose.[7]
Ecology/Pathology
The natural habitat for Shigella species is the intestinal tract of humans or other primates. They can live outside of the body when they are passed with the feces just long enough to be transmitted into drinking water or food in order to return back to the intestine.
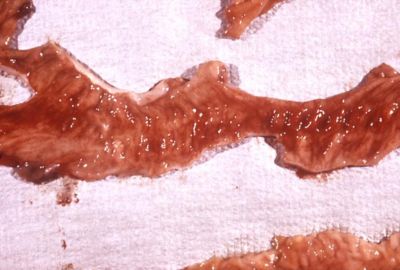
Shigella bacteria cause Shigellosis (bacillary dysentery) through oral-fecal transmission. Shigella is a highly infective agent able to infect a host with less than 20 organisms with an onset of about 12-48 hours, in favorable conditions. Once ingested, the Shigella makes its way through the gastrointestinal tract until it reaches the epithelial cells of the colonic intestinal mucosa, there it infects, causing irritation, inflammation and destruction. General symptoms include stomach cramps, high fever, mucus in feces, and bloody diarrhea due to ulceration of colonic lining and rectum. In most cases these symptoms are mild and resolve in about a week but other cases can become severe enough to be fatal without proper medical care. The elderly, very young and those weakened by disease are much more sensitive to the bacteria. In very young children very high fever may be accompanied with seizures.[4] Usually Shigella bacteria is found in areas of poor sanitation. Food washed with contaminated water or not cleaned properly may also be a target. In 1998 an outbreak of Shigellosis occurred in Chicago due to Shigella boydii type 18 found on the cilantro and parsley in bean salad.[6] Less than 10% of foodborne illnesses are attributed to Shigella bacteria in the United States. With an estimated 300,000 yearly incidents of shigellosis in the U.S. the percentage due to food contamination is unknown due to the difficulty in isolation. Given its infectiousness the number is undoubtedly high.[2] Around the world Shigella boydii is mainly restricted to the sub-continent of India.[3]
Current Research
1. Comparisons between Shigella species and between E. coli have been made and are being made in order to better understand Shigella's pathogenicity, patterns of infection, and varying harmfullness. Answers to these questions will hopefully lead to better avoidance and treatment of shigellosis.[3]
2. At the National Shigella Reference Centre in Israel during the years 2000- 2004 5,616 Shigella isolates were tested for resistance to certain antibacterial products. In one strain of Shigella boydii 2 and in two strains each of Shigella flexneri 2a, S. flexneri 6, and Shigella sonnei there was discovered resistance to ceftriaxone. All of these strains were found to be producers of extended-spectrum beta-lactamase (ESBL) and also very sensitive to tazobactam which inhibits the bacterial beta-lactamases. More research is needed to determine the rate of resistance to antibiotics and rate of transfer of resistant genes.[9]
3. At the French National Reference Centre for Escherichia coli and Shigella, from 2000- 2004, 4,198 Shigella isolates were obtained. 180 of those isolates, taken from patients infected with diarrhea and dysentery, did not respond to any available antisera (serum containing antibodies) that had been used on all known S. boydii serotypes. All of the isolates shared characteristics of S. boydii serotypes except for their rRNA genes were unique from all others. A new antiserum was manufactured and destroyed the new serotype S. boydii 20. 91 of the 180 patients had traveled internationally therefore further studies must be done in order to determine the epidemiology of this serogroup and the its potential widespread threat.[10]
References
1. Feng Lu, Sof'ya N. Senchenkova, Jinghua Yang, Alexander S. Shashkov, Jiang Tao, Hongjie Guo, Guang Zhao, Yuriy A. Knirel, Peter Reeves, and Lei Wang. "Structural and Genetic Characterization of the Shigella Boydii Type 13 O Antigen." J Bacteriol 186(2) (2004): 383-392. NCBI. PMID 14702307
2. "Shigella." U.S. Food and Drug Administration. 14 June 2006. Department of Health and Human Services. 30 Apr. 2007 <http://www.cfsan.fda.gov/~mow/chap19.html>.
3. Yang, Fan, Jian Yang, Xiaobing Zhang, and Yan Jiang. "Genome Dynamics and Diversity of Shigella Species, the Etiologic Agents of Bacillary Dysentery." Nucleic Acids Research 33 (2005): 6445-6458. Oxford Journals. <http://nar.oxfordjournals.org/cgi/content/abstract/33/19/6445?maxtoshow=&HITS=10&hits=10&RESULTFORMAT=&fulltext=shigella+boydii&searchid=1&FIRSTINDEX=0&resourcetype=HWCIT>.
4. "Shigellosis." CDC. 13 Oct. 2005. HHS. 1 May 2007 <http://www.cdc.gov/ncidod/dbmd/diseaseinfo/shigellosis_g.htm#What%20sort%20of%20germ%20is%20Shigella>.
5. Woodward, David L., Clifford G. Clark, Richard A. Caldeira, and Rafiq Ahmed. "Identification and Characterization of Shigella Boydii 20 Serovar Nov., a New and Emerging Shigella Serotype." Journal of Medical Microbiology 54 (2005): 741-748. Society of General Microbiology. <http://jmm.sgmjournals.org/cgi/content/abstract/54/8/741>.
6. Chan, Yc, and Hp Blaschek. "Comparative Analysis of Shigella Boydii 18 Foodborne Outbreak Isolate and Related Enteric Bacteria: Role of RpoS and AdiA in Acid Stress Response." NCBI 68 (2005): 521-7. NCBI. <http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=PubMed&cmd=Retrieve&list_uids=15771176&dopt=Citation>. PMID: 15771176
7. Holt, J. G., Krieg, N. R., Sneath, P. H. A., Staley, J. T. & Williams, S. T. (editors) (1994). Bergey’s Manual of Determinative Bacteriology, 9th edn. Baltimore, MD: Williams & Wilkins.
8. Ja, Giron. "Expression of Flagella and Motility by Shigella." Molecular Microbiology 18 (1995): 63-75. NCBI. <http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&list_uids=8596461&dopt=Abstract>. PMID: 8596461
9. Vasilev, V, R Japheth, R Yishai, N Andorn, L Valinsky, S Navon-Venezia, I, Chmelnitsky, Y Carmeli, and D Cohen. "Extended-Spectrum Beta-Lactamase-Producing Shigella Strains in Israel, 2000-2004." European Journal of Clinical Microbiology and Infectious Diseases 26 (2007): 189-194. NCBI. <http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=pubmed&cmd=Retrieve&dopt=AbstractPlus&list_uids=17265070&query_hl=4&itool=pubmed_docsum>.
10. Grimont,Francine, Monique Lejay-Collin, Kaisar A. Talukder, Isabelle Carle, Sylvie Issenhuth, Karine Le Roux1 and Patrick A. D. Grimont. "Identification of a group of shigella-like isolates as Shigella boydii 20." Jounal of Medical Microbiology 56 (2007): 749-754. Society for General Microbiology. <http://jmm.sgmjournals.org/cgi/content/abstract/56/6/749>.
11. S. boydii (serotype 4) Sb227, 4519823 bp, NC_007613 NCBI. <http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=genome&cmd=Retrieve&dopt=Overview&list_uids=19003>.
12. S. boydii (serotype 4) Sb227 pSB4_227, 126697 bp, NC_007608 NCBI. <http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=genome&cmd=Retrieve&dopt=Overview&list_uids=19008>.
Edited by James Cunningham a student of Rachel Larsen and Kit Pogliano