Oil-Eating Bacteria: A Tool for Bioremediation
Introduction
The use of petroleum, or crude oil, has been increasing since the industrial revolution [1]. The two common uses of petroleum are fuel and the synthesis of organic compounds [2]. Petroleum fuel is used as an energy source to provide many common-day luxuries, at least in the first world, such as heating and lighting for buildings and homes and for transportation [2]. Starting in 2000, petroleum was used to synthesize 99% of human-made organic compounds such as gasoline and kerosine [2]. Clearly, we rely heavily on petroleum and it is a ubiquitous chemical in our daily lives; however, there are a plethora of environmental consequences to such high petroleum consumption levels.

Petroleum pollution is one of the most common pollutants in the world and is harmful to almost every species on Earth [4]. Each year, three million tons of petroleum enters the ocean through activities such as oil-pumping, transportation, and more than half coming from prohibited activities and accidents [3]. Numerous studies have documented the negative consequences of petroleum pollution on organismal life. For example, petroleum has been found to interfere with the photosynthesis pathway of phytoplankton, which are the primary producers (base of the food chain) in marine environments [5]. Therefore, decreases in phytoplankton abundance caused by petroleum pollution could have bottom-up effects on the whole marine ecosystem [5]. Coral reefs are also negatively affected by petroleum, and a study by Turner and Renegar (2017) found that petroleum exposure can cause decreases in the abundance and diversity of coral by reducing growth rates, reproduction rates, and increasing tissue loss and death. Oil pollution has led to the death of many birds by coating their plumage and fur with oil which decreases their insulation and ability to fly ((Adams et al, 2015). Not only are marine environments affected by petroleum pollution, but terrestrial environments can also be affected through soil and groundwater contamination [4]. Clean groundwater trapped in aquifers is a precious resource for humans that provides most of our drinking water, making the contamination of groundwater a serious issue [4].
Given the multiple consequences of petroleum pollution today, it appears that we desperately need an environmentally-friendly technique to remove petroleum from contaminated ecosystems. One such technique involves bioremediation, which is the use of microbes to convert toxic pollutants, such as anthropogenic petroleum, into non-toxic compounds [6]. Recent scientific work has found that some bacteria (Oleispira, Oleiphilus, Thalassolituus, Alcanivorax and Cycloclasticus) can feed on petroleum due to their hydrocarbon‐degrading abilities [3]. Evolution has played a role in the formation of species capable of degrading petroleum, which is composed of stable hydrocarbons that are difficult to break down. Petroleum oil is naturally found in environments from seeps in the seafloor and from fatty acids and lipids produced by plants and animals [7]. To colonize these petroleum niches, some bacteria have evolved to produce enzymes capable of breaking down petroleum and using it as energy source for their metabolic pathways [7]. Therefore, there is hope in the scientific community that we can utilize oil-degrading bacteria and enhance their abilities through genetic engineering to create a bioremediation tool to remove anthropogenic petroleum from polluted environments.
Alcanivorax borkumensis
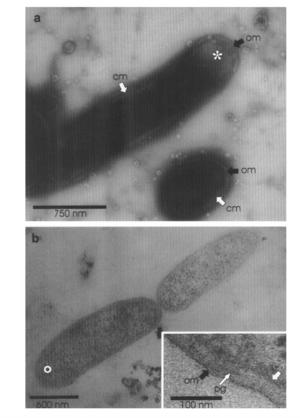
While multiple bacteria have the capacity to break down petroleum, Alcanivorax borkumensis appears to be the dominant bacteria among oil-degrading bacteria, making up 80% to 90% of the microbes in oil-polluted water [10]. A. borkumensis was first discovered in 1998 by a team of microbiologists led by Michail Yakimov [8]. The researchers isolated A. borkumensis from sea water and sediment samples in the North Sea, and through experimental analysis they found that the bacteria is a rod-shaped, gram-negative, non-spore-forming, non-motile, aerobic bacteria in the γ-proteobacterium phylum [8]. Since it was first discovered in 1998, A. borkumensis has been observed across the world, from the Pacific Ocean to the Arctic Sea [10]. The figure on the left is from electron microscopy imaging and shows A. borkumensis and its outer membrane [8], a requisite for gram-negative bacteria.
A. borkumensis grows slowly and can grow almost entirely on alkanes [10], which is one of the major compounds found in petroleum. In unpolluted water, A. borkumensis abundance is generally low; however, these bacteria become abundant in oil-polluted waters. For optimal growth, the bacteria requires a temperature between 25 and 30°C, but they can grow between 4 and 35°C [8]. Additionally, the necessary salinity concentration for A. borkumensis can be as low as 1% and as high as 12.5%, with the optimal salinity concentration ranging from 3 to 10% NaCl [8]. Yakimov and researchers found that A. borkumensis are 0.6-0.8 μm wide and 16-25 μm long if growing on pyruvate-supplemented medium, but are much shorter (1-1.5 μm) if n-alkanes are the sole carbon source. Additionally, the cell wall of A. borkumensis is generally 31 to 36 nm [8].
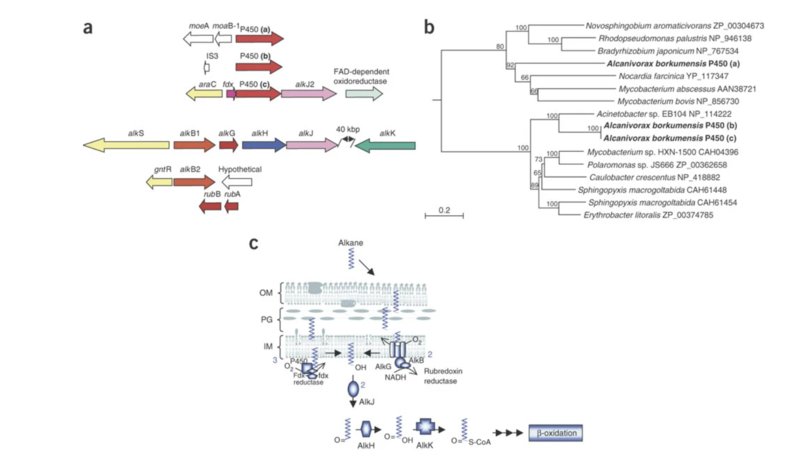
The key to A. borkumensis superior ability to degrade petroleum can be found in its genome, which contains a circular genome of 3,120,143 base pairs (bp) and a mean G+C content of 54.7% [10]. Within the genome of A. borkumensis are a plethora of different systems that help the bacteria attach itself to petroleum through pili and biosurfactants and degrade petroleum through enzyme formation [10]. For instance, A. borkumensis has 16 pili genes that help the bacteria attach themselves to oil in water. In addition, many secretion systems, such as the Type II secretion system, Sec translocon, twin-arginin translocation (Tat), and five genes from the HlyD family, are used by A. borkumensis to help them create biofilms, which are a cluster of microorganisms living on a surface [10]. After bacteria have attached themselves to the surface of petroleum, they need a way to degrade petroleum hydrocarbons. Within the genome of A. borkumensis is the alkSB1GHJ operon, which is known to contain genes for the alkane degradation pathway [10]. One important gene in this operon is the AlkB1 gene which makes an alkane hydroxylase that oxidizes medium-chain alkanes with 5 to 12 carbons (Schneiker et al., 2006). AlkB2 is another gene with the same function as AlkB1 but it hydrolyzes medium-chain alkanes with 8 to 16 carbons and it is encoded by a different alkane hydroxylase system (ABO_0122) [10]. By searching through databases, Schneiker and colleagues found that relatively few marine species have AlkB genes and even fewer have two types of this gene. In addition to the genes discussed above, A. borkumensis also has three cytochromes (ABO_2384 for P450, ABO_2288 P450, and ABO_0201 P450) that contain genes involved in alkane degradation [10]. Surprisingly, even more enzyme-encoding genes for alkane degradation, such as alkK, alkL, alkN, rubA, and rubB, were also found in the A. borkumensis genome [10]. Due to the wide range of alkane degradation pathways, A. borkumensis is able to efficiently break down petroleum compared to other species. The figure below shows the genes involved in alkane degradation (a) as well as a phylogenetic tree of bacteria with similar P450 cytochromes (b) and the possible alkane degradation pathway (c).
Other Bacteria Capable of Petroleum Degradation

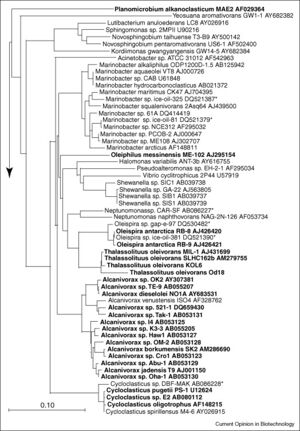
Besides A. borkumensis, there are a number of other bacteria, and even some fungi, that can degrade oil. Since the first hydrocarbon degrading bacteria was discovered around a century ago, scientists have found 200 genera of bacteria, cyanobacteria, algae, and fungi that can degrade hydrocarbons, which adds up to over 500 different species (Yakimov et al, 2007).
Marine hydrocarbon-degrading microbes tend to be specialized to consume hydrocarbons and are often referred to as obligate hydrocarbonoclastic bacteria (OHCB) (Yakimov et al, 2007). The five genera of OHCBs include Oleispira, Oleiphilus, Thalassolituus, Alcanivorax and Cycloclasticus, and the figure to the left shows where the 16S rRNA of these different genera have been found so far (Yakimov et al, 2007). OHCBs play an important role in the global carbon cycle by breaking down stable hydrocarbons that most other species are unable to degrade (Yakimov et al, 2007). Mesocosm studies have found that when oil is inserted into marine water there is a sudden shift in the microbial community composition (Yakimov et al, 2007). One study conducted in Sicily found that the microbial community of unpolluted water contains mostly Vibrio, Arcobacter, and some gram-positive bacteria, but after the addition of petroleum the microbial community shifted to one made up of mostly HCBs, with Alcanivorax being the most abundant (Yakimov et al, 2007). While Alcanivorax species are usually the first to increase in abundance, they are followed by Cycloclasticus spp. that can degrade some of the more difficult petroleum compounds like aromatic compounds (Kasai et al., 2002). Other studies have added temperature as a variable affecting hydrocarbon degradation, and researchers have found that at low temperatures (4°C) the genus Oleispira dominates (Yakimov et al, 2007). Oleispira species are adapted to the cold and have been found in Antarctic seawater (Yakimov et al, 2007), which could make them a useful bioremediation tool in colder climates. Oleiphilus species and their hydrocarbon abilities have been less studied (Yakimov et al, 2007). However, these bacteria are believed to be sponge symbionts because they have mostly been isolated from sponges in the North Atlantic and equator region in the Pacific Ocean (Yakimov et al, 2007).
While this page is mainly about oil-eating bacteria, it is important to note that petroleum degradation is not only limited to bacteria but can be accomplished by some fungi. So far 103 fungi genera are known to have petroleum degradation abilities (Xue et al, 2015). Some of the most common hydrocarbon degrading fungi include Ascomycota, Basidiomycota, and Mucoromycotina (Simister et al., 2015). Fungi are able to break down multiple oil compounds due to their diverse enzymes (Simister et al., 2015). Enzymes used by fungi that can degrade hydrocarbons include both intracellular compounds (cytochrome P450 monooxygenases, nitroreductases and transferases) and extracellular enzymes (laccases and fungal peroxidases) (Simister et al., 2015). A study by Hasan AI- Jawhari (2014) found that one species of fungi, Aspergillus niger, was able to degrade 95% of the petroleum it was given after only 28 days. Due to the ability of both bacteria and fungi to degrade petroleum, some scientists have looked into the effect of bacteria-fungi consortiums as a tool for bioremediation due to possible co-metabolism pathways that could increase degradation (Ghorbannezhad et al, 2018). Fungal-bacterial mixed cultures have been observed to degrade petroleum hydrocarbons more efficiently than bacteria and fungi separately, which indicate that mixed cultures should be considered as one means of petroleum bioremediation (Ghorbannezhad et al, 2018).
Biodegradation Mechanism and Pathway
The mechanism and pathway through which bacteria degrade petroleum hydrocarbons is important to understand if we want to utilize this mechanism for bioremediation. Petroleum degradation can occur anaerobically and aerobically, but it commonly occurs aerobically, meaning oxygen is used as the terminal electron acceptor (Nilanjana and Chandran, 2011). The four main types of hydrocarbons are saturates, aromatics, asphaltenes, and resins (Nilanjana and Chandran, 2011). Hydrocarbons are composed mainly of hydrogens and carbons and lack functional groups, which makes them unreactive at room temperatures (Victor et al, 2020). Therefore, hydrocarbon-degrading bacteria must use a pathway that allows them to break down these exceptionally stable compounds (Nilanjana and Chandran, 2011).
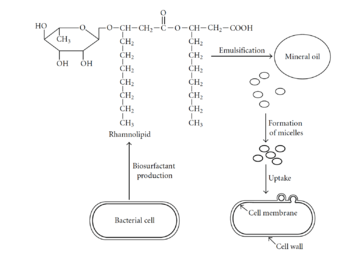
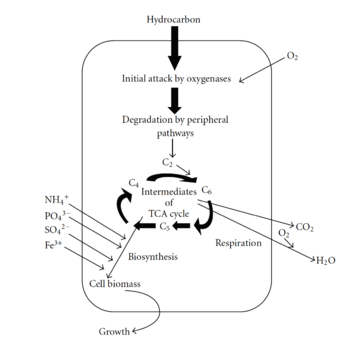
The first step of the degradation process involves hydrocarbon uptake through the use of biosurfactants (Nilanjana and Chandran, 2011). Biosurfactants help microbes attach to petroleum hydrocarbons by decreasing the surface tension and increasing the petroleum surface area available to bacteria (Nilanjana and Chandran, 2011). Additionally, biosurfactants aid in the formation of micelles, which is a sphere-shaped ball of petroleum that can get incorporated into the bacterial cell (Nilanjana and Chandran, 2011). Once petroleum enters the bacterial cell, it must be degraded by enzymes. The first enzymes involved in degradation are oxygenases and peroxidases, which also help bring oxygen into the cell (Nilanjana and Chandran, 2011). The next step in the pathway is called the peripheral degradation pathway that takes the organic pollutants and converts them into metabolic intermediates (Nilanjana and Chandran, 2011). Some of these intermediates can then enter the tricarboxylic acid cycle, and be used for biosynthesis and cell growth (Nilanjana and Chandran, 2011).
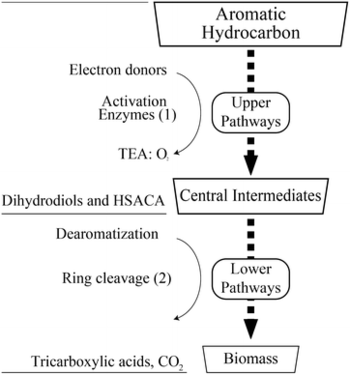
To take a more in depth look at the biodegradation mechanism, we will focus on aromatic hydrocarbons specifically, because they are one of the more difficult petroleum hydrocarbons to degrade (Ladino-Orjuela et al, 2016). Aromatic hydrocarbons contain a benzene ring, which is very chemically stable due to its pi bonds. Degradation of aromatic is broken down into two steps: the upper pathway and the lower pathway with central intermediates in-between these two pathways (Ladino-Orjuela et al, 2016). To start the upper pathway process, an enzyme called monooxygenases splits oxygen gas into two molecules and places one of the oxygens into the benzene ring while reducing the other oxygen atom to water (Ladino-Orjuela et al, 2016). This reaction results in the formation of phenol, which then gets converted to catechol through the addition of another oxygen molecule by phenol hydroxlase (Ladino-Orjuela et al, 2016). Catechol is one of the main central intermediate in the aromatic degradation process; however, there are a number of other intermediates such as hydroquinone, (chloro)hydroxyquinol, hydroxyhydroquinone, hydroxy-substituted aromatic, and carboxylic acids (Ladino-Orjuela et al, 2016). These intermediates can then enter the lower pathway where they undergo dearomatization followed by ring cleavage (Ladino-Orjuela et al, 2016). Ring cleavage is accomplished by multiple dioxygenases enzymes and these cleavages result in tricarboxylic acids. Tricarboxylic acids enter the TCA cycle and this completes the degradation process (Ladino-Orjuela et al, 2016).
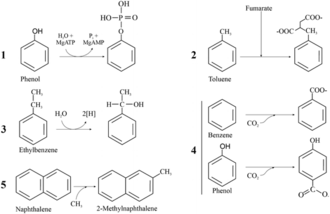
Aromatic degradation can also occur under anaerobic conditions. This is important because there are lots of environments that do not contain oxygen and can become polluted, such as deep parts of the ocean and soil (textbook). There are currently five know ways for the activation of aromatic hydrocarbons degradation (upper pathway) in anaerobic conditions: phosphorylation, fumarate insertion, O2-independente hydroxylation, carboxylation, and methylation (see the figure to the right for a visual depiction) (Ladino-Orjuela et al, 2016). The most common central intermediate under anaerobic conditions is benzoyl-CoA (Ladino-Orjuela et al, 2016). Other intermediates include 3-methyl-benzoyl-CoA, 3-hydroxy-benzoyl-CoA, 2-amino-benzoyl-CoA, phloroglucinol, resorcinol, and hydroxyhydroquinone (Ladino-Orjuela et al, 2016). Benzoyl-CoA enters the lower pathway by benzoyl-CoA reductase causing dearomatization (Ladino-Orjuela et al, 2016). Ring cleavage of benzoyl-CoA is accomplished by 6-oxo-2-hydroxycyclo-hexane-1-carboxyl-CoA hydrolase that forms 3-hydroxypimelyl-CoA, which enters the TCA cycle (Ladino-Orjuela et al, 2016).
Many factors can affect biodegradation and the rate at which it occurs. For instance, oxygen availability is an important factor because degradation is often faster in aerobic environments compared to anaerobic (Victor et al, 2020). Temperature is another factor that affects the rate of biodegradation with lower temperatures causing slower degradation (Victor et al, 2020). Additionally, different environments have different maximum degradation rates: soils occur at 30 to 40°C, freshwater at 20 to 30°C, and marine at 15 to 20°C (Victor et al, 2020). Nutrient levels are another key factor affecting degradation rates. Studies have found that when oil spills occur in aquatic environments nitrogen and phosphorus levels soon become the limiting factor affecting petroleum degradation (Victor et al, 2020). Therefore, nutrient additions will likely be needed for bioremediation. An additional factor is light, which often increases the degradation rates of hydrocarbons by direct photochemical action (Victor et al, 2020). High salinity often decreases or inhibits degradation, but its effect on degradation can vary in different environments and between bacteria species (Victor et al, 2020).
[13]
The Bioremediation Tool
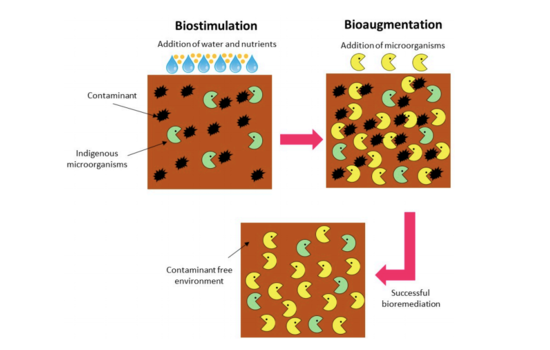
Bioremediation is the most cost-effective and environmentally friendly means of restoring an environment contaminated with petroleum [16]. Since petroleum is biodegradable, it will naturally be metabolized by microbes and converted into non-toxic compounds such as CO2, CH4, H2O and biomass [16]
. Microbial bioremediation is a technique that can help speed up this process [16]. The two methods for increasing the rate of biodegradation are bioaugmentation and biostimulation. Bioaugmentation is when hydrocarbon-degrading microbes are added to polluted waters or soil, and biostimulation is when environmental variables, such as the amount of limiting nutrients, are altered to “stimulate” the native hydrocarbon-degrading microbes [17]. These two methods can occur either in situ or ex situ [16]. In situ bioremediation is when the polluted material is treated at the site, and ex situ is when the pollutant is extracted from the site and treated in a different location [16]. Ex situ bioremediation is generally more expensive [16].
One means of biostimulation that has been well tested is the use of water‐soluble nutrient products [17]. In the field, nutrients are sprayed onto the polluted area or dry granules containing nutrients are dispersed [17]. Multiple field trials have found this to be an effective method for increasing petroleum degradation (Venosa et al, 1996, Swannell et al, 1996, and Roling et al, 2004). However, this method does come with limitations as water soluble nutrients can easily get washed out of an environment and cause eutrophication [17]. If this method is to be utilized, there needs to be an effective way for nutrients to be contained in specific locations. One way to do this is through slow-release fertilizers, which consist of a solid inorganic form of nutrients wrapped in hydrophobic compounds [17]. A study by Kasai et al. (2004) found that the addition of slow-release fertilizers caused the degradation of more than 90% of n‐alkanes, 60% of (alkyl)naphthalenes and between 30% and 40% of aromatics over 30 days. However, some studies have found that slow-release fertilizers do not cause a significant difference compared to unfertilized areas and are therefore ineffective at stimulating degradation [17]. Because of this, more work needs to be done to figure out techniques that would increase nutrients at a rate fast enough to stimulate microbes but slow enough that all the nutrients do not get washed out [17]. In addition to altering nutrient levels, biostimulation also can occur through adjusting oxygen levels, temperature, and the pH of a contaminated area
[18].
Bioaugmentation was first introduced in the late 1960s by George Robinson, who added microbes to an area in Santa Barbara, California, where an oil spill had occurred [18]. Since then, multiple species have been tested and utilized for bioremediation [18]. The best microbes for bioaugmentation are ones that can survive in multiple environmental conditions and can degrade the majority of the compounds that make up petroleum (Zaida and Piakong, 2018). For those reasons, A. borkumensis is one of the best known petroleum-degrading microbes. A study by Hassanshahian et al. (2014) found that bioaugmentation with A. borkumensis degraded petroleum in an artificially polluted mesocosm by 95% over 20 days. Another way to go about bioaugmentation is through microbial consortiums, which are a mixture of bacteria cultures. Microbial consortiums have been found to increase petroleum degradation by co-metabolism that results in enhanced biodegradation. Co-metabolism allows the intermediates produced by one species to be degraded by another species that might contain the appropriate metabolic pathway missing from the first bacteria [19]. A bacterial consortium designed by Bento et al. (2005) found that the consortium was able to degrade petroleum at a 1.5-fold higher rate than single bacterial strains alone [19]. Therefore, microbial consortia are a promising means of petroleum bioremediation.
To enhance bioaugmentation, some studies have looked into the effects of genetically engineered hydrocarbon-degrading bacteria [18]. . Recombinant DNA techniques can be used to insert genes into microorganisms and make them more efficient at breaking down petroleum [18]. For example, one study conducted at the University of Tennessee inserted a plasmid containing genes that allowed for naphthalene (a type of hydrocarbon) catabolism into the bacteria Pseudomonas fluorescens [18]. After insertion, there was an increase in naphthalene degradation [18]. Genetically engineered bacteria can be an extremely powerful tool for increasing the effectiveness of biodegradation [18]. Engineering of microbes can allow different degradation pathways to be combined, leading to complete petroleum breakdown [18]. Currently, the main limitation on genetically engineered bacteria is government regulations. While keeping public safety in mind is important, the approval process for genetically engineered organisms can be long and often includes many refinement stages that make the road to commercial development a strenuous one [18].
Conclusion
Oil-eating bacteria appear to be one of the most robust and ecologically friendly ways to break down petroleum. As more and more petroleum is making its way into Earth’s oceans and soils, continued research on hydrocarbon-degrading microbes is particularly important. Future research is needed to establish effective consortiums of bacteria that can be utilized in multiple environmental conditions. Additionally, genetically engineered bacteria have potential as bioremediation tools. There are a plethora of different pathways for hydrocarbon degradation, with many of the enzymes and metabolic participants still a mystery [20] . To increase our chances of restoring the numerous environments around the world that are affected by petroleum pollution, additional studies are needed to broaden and deepen our understanding.
References
- ↑ Villela, H.D.M, Peixoto, R.S., Soriano, A.U., Carmo, F.L. (2019). Microbial bioremediation of oil contaminated seawater: A survey of patent deposits and the characterization of the top genera applied. Science of The Total Environment. 666: 743-758.
- ↑ 2.0 2.1 2.2 Eneh, O. C. (2011). A Review on Petroleum: Source, Uses, Processing, Products and the Environment. Journal of Applied Sciences. 11(12): 2084-2091.
- ↑ 3.0 3.1 3.2 Brooijmans, R. J., Pastink, M. I., & Siezen, R. J. (2009). Hydrocarbon-degrading bacteria: the oil-spill clean-up crew. Microbial biotechnology, 2(6), 587–594
- ↑ 4.0 4.1 4.2 Logeshwaran, P., Megharaj, M., Chadalavada, S., Bowman, M., Naidu, R.. (2018). Petroleum hydrocarbons (PH) in groundwater aquifers: An overview of environmental fate, toxicity, microbial degradation and risk-based remediation approaches. Environmental Technology & Innovation. 10: 175-193.
- ↑ 5.0 5.1 Jiang, Z., Huang, Y., Xu, X., Liao, Y., Shou, L., Liu, J., Chen, Q., Zeng, J. (2010).Advance in the toxic effects of petroleum water accommodated fraction on marine Plankton. Acta Ecologica Sinica. 30(1): 8-15.
- ↑ Ezezika, O.C. and Singer, P.A. (2010). Genetically engineered oil-eating microbes for bioremediation: Prospects and regulatory challenges. Technology in Society. 32(4): 331-335.
- ↑ 7.0 7.1 Yakimov, M.M. Timmis, K.N., Golyshin, P.N. (2007). Obligate oil-degrading marine bacteria. Current Opinion in Biotechnology. 18(3): 257-266.
- ↑ 8.0 8.1 8.2 8.3 8.4 8.5 8.6 Yakimov, Michail M.; et al. (1998). "Alcanivorax Borkumensis gen. nov., sp. nov., A New, Hydrocarbon-degrading And Surfactant-producing Marine Bacterium". International Journal of Systematic Bacteriology. 48 (2): 339–348
- ↑ Julia S. Sabirova, Anke Becker, Heinrich Lünsdorf, Jean-Marc Nicaud, Kenneth N. Timmis, Peter N. Golyshin. (2011). Transcriptional profiling of the marine oil-degrading bacterium Alcanivorax borkumensis during growth on n-alkanes, FEMS Microbiology Letters. 319(2): 160–168
- ↑ 10.00 10.01 10.02 10.03 10.04 10.05 10.06 10.07 10.08 10.09 10.10 Schneiker, S., dos Santos, V., Bartels, D. et al. (2006) Genome sequence of the ubiquitous hydrocarbon-degrading marine bacterium Alcanivorax borkumensis. Nat Biotechnol 24, 997–1004.
- ↑ 11.0 11.1 Yakimov, M.M., Timmis, K.N., Golyshin, P.N. (2007). Obligate oil-degrading marine bacteria. Current Opinion in Biotechnology. 18(3): 257-266.
- ↑ Nur Zaida Z., Piakong M.T. (2018) Bioaugmentation of Petroleum Hydrocarbon in Contaminated Soil: A Review. In: Kumar V., Kumar M., Prasad R. (eds) Microbial Action on Hydrocarbons. Springer, Singapore.
- ↑ 13.0 13.1 13.2 Victor, I., Iwok, E., Archibong, I., Effiom, O., Okon, E., Andem, A. (2020). The biochemical mechanisms of petroleum degradation by bacteria. International Journal of Scientific and Engineering Research. 11. 1258-1275.
- ↑ 14.0 14.1 Ladino-Orjuela G., Gomes E., da Silva R., Salt C., Parsons J.R. (2016) Metabolic Pathways for Degradation of Aromatic Hydrocarbons by Bacteria. In: de Voogt W. (eds). Reviews of Environmental Contamination and Toxicology. 237. Reviews of Environmental Contamination and Toxicology (Continuation of Residue Reviews), vol 237. Springer, Cham.
- ↑ Silva, Israel & Almeida, Fabíola & Silva, Nathália & Casazza, Alessandro & Converti, Attilio & Sarubbo, Leonie. (2020). Soil Bioremediation: Overview of Technologies and Trends. Energies. 13. 4664.
- ↑ 16.0 16.1 16.2 16.3 16.4 16.5 Yuniati, M.D. (2017) Bioremediation of petroleum-contaminated soil: A Review. IOP Conf. Series: Earth and Environmental Science 118: 012063
- ↑ 17.0 17.1 17.2 17.3 17.4 17.5 17.6 Nikolopoulou, M. and Kalogerakis, N. (2009). Biostimulation strategies for fresh and chronically polluted marine environments with petroleum hydrocarbons. Journal of Chemical Technology and Biotechnology. 84: 802-807.
- ↑ 18.0 18.1 18.2 18.3 18.4 18.5 18.6 18.7 18.8 18.9 Adams, G.O., Fufeyin, P.T., Okoro, S.E., Ehinomen, I. (2015). Bioremediation, Biostimulation and Bioaugmentation: A Review. International Journal of Environmental Bioremediation & Biodegradation. 3(1): 28-39.
- ↑ 19.0 19.1 [ https://doi.org/10.1007/978-981-13-1840-5_17 Zaida N.Z., Piakong M.T. (2018) Bioaugmentation of Petroleum Hydrocarbon in Contaminated Soil: A Review. In: Kumar V., Kumar M., Prasad R. (eds) Microbial Action on Hydrocarbons. Springer, Singapore. ]
- ↑ Ladino-Orjuela G., Gomes E., da Silva R., Salt C., Parsons J.R. (2016) Metabolic Pathways for Degradation of Aromatic Hydrocarbons by Bacteria. In: de Voogt W. (eds). Reviews of Environmental Contamination and Toxicology. 237. Reviews of Environmental Contamination and Toxicology (Continuation of Residue Reviews), vol 237. Springer, Cham.
Authored for BIOL 238 Microbiology, taught by Joan Slonczewski, 2021, Kenyon College.