Streptococcus parasanguinis and the Development of Dental Plaque
Introduction
Select a topic about genetics or evolution in a specific organism or ecosystem.
Overall text length (all text sections) should be at least 1,000 words (before counting references), with at least 2 images.
The topic must include one section about microbes (bacteria, viruses, fungi, or protists). This is easy because all organisms and ecosystems have microbes.
Compose a title for your page.
Type your exact title in the Search window, then press Go. The MicrobeWiki will invite you to create a new page with this title.
Open the BIOL 116 Class 2024 template page in "edit."
Copy ALL the text from the edit window.
Then go to YOUR OWN page; edit tab. PASTE into your own page, and edit.

At right is a sample image insertion. It works for any image uploaded anywhere to MicrobeWiki. The insertion code consists of:
Double brackets: [[
Filename: PHIL_1181_lores.jpg
Thumbnail status: |thumb|
Pixel size: |300px|
Placement on page: |right|
Legend/credit: Electron micrograph of the Ebola Zaire virus. This was the first photo ever taken of the virus, on 10/13/1976. By Dr. F.A. Murphy, now at U.C. Davis, then at the CDC.
Closed double brackets: ]]
Other examples:
Bold
Italic
Subscript: H2O
Superscript: Fe3+
Genetics
Streptococcus parasanguinis (S. parasanguinis) is a gram-positive bacterium commonly found in the human oral cavity, particularly in the dental plaque of healthy individuals. Its genome offers valuable insights into its adaptability and role in dental plaque formation. Through genomic studies, scientists have uncovered key features that enable S. parasanguinis to thrive in the oral environment and contribute to oral health.
The bacterium's genome reveals a versatile genetic makeup that allows it to metabolize a wide range of carbohydrates. This includes simple sugars like glucose and more complex carbohydrates, such as glycans found in the salivary pellicle on tooth surfaces. These capabilities are crucial for the bacterium’s survival in the fluctuating nutrient environment of the oral cavity. Through fermentation, S. parasanguinis produces acids, which lower the pH of its surroundings, potentially making them more favorable for the growth of other oral microbes (Lynch et al., 2015). This metabolic flexibility supports its role in biofilm formation, which is essential in the early stages of dental plaque development.
A critical aspect of S. parasanguinis is its ability to form strong attachments to tooth surfaces and other microorganisms, facilitated by genetic elements encoding adhesins. Adhesins are surface proteins that mediate bacterial adhesion to host tissues and microbial communities. Among these adhesins, the SspA protein stands out for its role in binding to host proteins and extracellular matrix components, which enhances the bacterium’s ability to anchor itself to tooth and mucosal surfaces. This interaction is essential for the establishment of microbial communities that form the foundation of dental plaque (Kreth et al., 2012).
Moreover, S. parasanguinis possesses genetic pathways that enable it to survive in the dynamic oxygen conditions of the oral cavity. The oral environment is marked by varying levels of oxygen, from aerobic conditions on the surface of biofilms to anaerobic conditions in deeper layers. S. parasanguinis contains genes that help it withstand oxidative stress, allowing it to persist in both oxygen-rich and oxygen-deprived areas of the biofilm (Morrow et al., 2019). This genetic adaptability contributes to its resilience within the oral ecosystem.
The bacterium also produces extracellular matrix components, such as polysaccharides, which form the biofilm’s scaffold. These substances provide structural integrity to the biofilm and protect the bacterial community from host immune responses and antimicrobial treatments. The production of these extracellular substances further supports S. parasanguinis in maintaining its position in the oral cavity (Kreth et al., 2012).
In addition to its biofilm-forming capabilities, S. parasanguinis plays a significant role in oral health and disease. The bacterium is involved in both the development of dental plaque and its potential role in oral diseases like periodontitis. Comparative genomic studies have shown that S. parasanguinis shares genetic similarities with other oral streptococci, such as Streptococcus gordonii, particularly in genes related to cell wall biosynthesis, stress responses, and metabolic pathways (Lynch et al., 2015). The genetic regulation of virulence factors in S. parasanguinis, such as those involved in biofilm formation, is influenced by environmental factors like oxygen levels, pH, and nutrient availability. This capacity for adaptation to changing conditions highlights the bacterium’s role in both maintaining oral health and potentially contributing to disease when the balance of the oral microbiome is disturbed.
Furthermore, S. parasanguinis has been shown to engage in horizontal gene transfer, a process that allows it to acquire genetic material from other bacteria. This mechanism can contribute to genetic diversity and the development of antibiotic resistance, further enhancing its ability to survive in the competitive oral environment (Wang et al., 2012). The bacterium also contains genes that encode immunoglobulin A (IgA) proteases, which may assist in immune modulation during chronic infections, suggesting its involvement in immune system interactions (Morrow et al., 2019).
Section titles are optional.
Include some current research, with at least one image.
Call out each figure by number (Fig. 1).
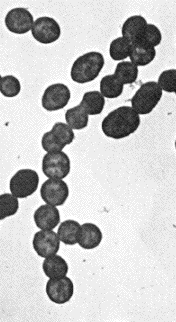
At right is a sample image insertion. It works for any image uploaded anywhere to MicrobeWiki. The insertion code consists of:
Double brackets: [[
Filename: pic1.gif
Thumbnail status: |thumb|
Pixel size: |300px|
Placement on page: |right|
Legend/credit: Gram-staining showing Gram-positive uniformly stained S. parasanguinis. seen under 1000× magnification.
Closed double brackets: ]]
Sample citations: [1]
[2]
A citation code consists of a hyperlinked reference within "ref" begin and end codes.
For multiple use of the same inline citation or footnote, you can use the named references feature, choosing a name to identify the inline citation, and typing [4]
Second citation of Ref 1: [1]
Here we cite April Murphy's paper on microbiomes of the Kokosing river. [5]
Section 2 Biome
Include some current research, with a second image.
Here we cite Murphy's microbiome research again.[5]
Section 3 Forming Dental Plaque
Conclusion
You may have a short concluding section.
Overall, cite at least 5 references under References section.
References
Edited by Amelia Russell, student of Joan Slonczewski for BIOL 116, 2024, Kenyon College.
- ↑ 1.0 1.1 Hodgkin, J. and Partridge, F.A. "Caenorhabditis elegans meets microsporidia: the nematode killers from Paris." 2008. PLoS Biology 6:2634-2637.
- ↑ Bartlett et al.: Oncolytic viruses as therapeutic cancer vaccines. Molecular Cancer 2013 12:103.
- ↑ Lee G, Low RI, Amsterdam EA, Demaria AN, Huber PW, Mason DT. Hemodynamic effects of morphine and nalbuphine in acute myocardial infarction. Clinical Pharmacology & Therapeutics. 1981 May;29(5):576-81.
- ↑ 4.0 4.1 text of the citation
- ↑ 5.0 5.1 Murphy A, Barich D, Fennessy MS, Slonczewski JL. An Ohio State Scenic River Shows Elevated Antibiotic Resistance Genes, Including Acinetobacter Tetracycline and Macrolide Resistance, Downstream of Wastewater Treatment Plant Effluent. Microbiology Spectrum. 2021 Sep 1;9(2):e00941-21.
